













| General Information: |
|
| Name(s) found: |
CG4199-PI /
FBpp0291627
[FlyBase]
CG4199-PH / FBpp0289483 [FlyBase] CG4199-PG / FBpp0289482 [FlyBase] CG4199-PB / FBpp0070322 [FlyBase] CG4199-PD / FBpp0070321 [FlyBase] CG4199-PC / FBpp0070320 [FlyBase] |
| Description(s) found:
Found 71 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 593 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
cell redox homeostasis
[IEA]
oxidation reduction [IEA] |
| Molecular Function: |
FAD binding
[IEA]
oxidoreductase activity [IEA] 2 iron, 2 sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSVNCKEYK TTGSTPKKEK STGGAVGSYQ SKCSNQSPST TMSTEESSPD SEYTSAVPVD 60
61 CRVTDLKENE MKQVDFDEDT RVLLVKQNDR LLAVGAKCTH YGAPLQTGAL GLGRVRCPWH 120
121 GACFNLENGD IEDFPGLDSL PCYRVEVGNE GQVMLRAKRS DLVNNKRLKN MVRRKPDDQR 180
181 VFIVVGGGPS GAVAVETIRQ EGFTGRLIFV CREDYLPYDR VKISKAMNLE IEQLRFRDEE 240
241 FYKEYDIELW QGVAAEKLDT AQKELHCSNG YVVKYDKIYL ATGCSAFRPP IPGVNLENVR 300
301 TVRELADTKA ILASITPESR VVCLGSSFIA LEAAAGLVSK VQSVTVVGRE NVPLKAAFGA 360
361 EIGQRVLQLF EDNKVVMRME SGIAEIVGNE DGKVSEVVLV DDTRLPCDLL ILGTGSKLNT 420
421 QFLAKSGVKV NRNGSVDVTD FLESNVPDVY VGGDIANAHI HGLAHDRVNI GHYQLAQYHG 480
481 RVAAINMCGG VKKLEAVPFF FTLIFGKGIR YAGHGSYKDV IIDGSMEDFK FVAYFINEAD 540
541 TVTAVASCGR DPIVAQFAEL ISQGKCLGRG QIEDPATRED WTKKLGQPLP QVR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
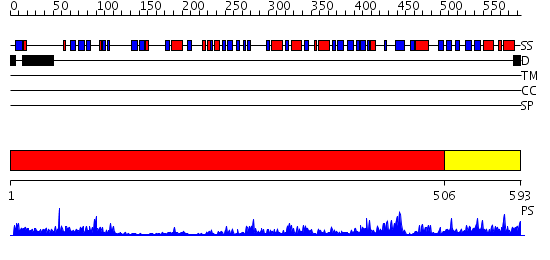
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..505] | 14.0 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 2 | View Details | [506..593] | 62.30103 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)