













| General Information: |
|
| Name(s) found: |
FBpp0301951
[FlyBase]
mod-PA / FBpp0085233 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
cytoplasm [IDA] protein complex [IPI] nucleolus [IDA] |
| Biological Process: |
cell proliferation
[IMP]
spermatid development [IMP] |
| Molecular Function: |
nucleotide binding
[IEA]
DNA binding [IDA][NAS] protein binding [IPI][NAS] mRNA binding [ISS][NAS] sequence-specific DNA binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQKKAVTVK GKKATNGEEK PLAKRVTKST KVQEEETVVP QSPSKKSRKQ PVKEVPQFSE 60
61 EDESDVEEQN DEQPGDDSDF ETEEAAGLID DEAEEDEEYN SDDEEDDDDD ELEPGEVSKS 120
121 EGADEVDESD DDEEAPVEKP VSKKSEKANS EKSEENRGIP KVKVGKIPLG TPKNQIVFVT 180
181 NLPNEYLHKD LVALFAKFGR LSALQRFTNL NGNKSVLIAF DTSTGAEAVL QAKPKALTLG 240
241 DNVLSVSQPR NKEENNERTV VVGLIGPNIT KDDLKTFFEK VAPVEAVTIS SNRLMPRAFV 300
301 RLASVDDIPK ALKLHSTELF SRFITVRRIS QESISRTSEL TLVVENVGKH ESYSSDALEK 360
361 IFKKFGDVEE IDVVCSKAVL AFVTFKQSDA ATKALAQLDG KTVNKFEWKL HRFERSTSGR 420
421 AILVTNLTSD ATEADLRKVF NDSGEIESII MLGQKAVVKF KDDEGFCKSF LANESIVNNA 480
481 PIFIEPNSLL KHRLLKKRLA IGQTRAPRKF QKDTKPNFGK KPFNKRPAQE NGGKSFVKRA 540
541 RF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
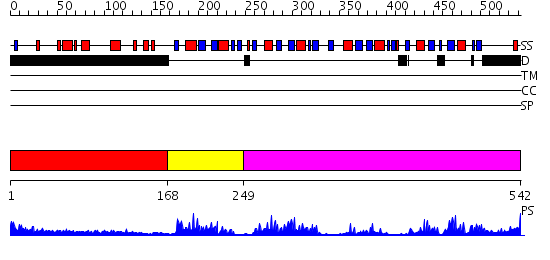
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 6.356996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [168..248] | 38.522879 | Poly(A)-binding protein | |
| 3 | View Details | [249..542] | 19.69897 | No description for 2ghpA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)