













| General Information: |
|
| Name(s) found: |
zwilch-PA /
FBpp0085138
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 641 amino acids |
Gene Ontology: |
|
| Cellular Component: |
kinetochore
[IDA]
|
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IMP]
mitosis [IMP] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASANLANV YAELMRRCGE SYTITYGAPP TYLVSMVGAA EAGKKIVLVF KEDRNGAVAR 60
61 LRTTPTRAAP KKEGSADLDL TGSPLKDDCL VDAIADLSID LQLDHSNPWK LEEEYQRGIP 120
121 VDKARSIVCS EFLQLAEGLG SVWFLCDGSD LGQTQLLQYE FNPTHFSRGI LSYQGVRPAY 180
181 LVTSQALVRH HGKTPDETLI ENSYQVNPHM RLRCSWTSSA ALPLLVNLND CDVALNHTFR 240
241 VGDCGPLTQD FMNQLRILVY IREDIVSYHT DVKQGVSRDP TYRCGSGIDM DELRESINQT 300
301 MTDVSGLIGR YSISNAEFDI EDVVQRAKVR QLTDLTDKLW ELLKCCHSYK DLKIAFSMLF 360
361 QCAARCNIVN TPTNKNRLAK IITELANRRL AMPCLSGAEP LELLLEIGLE KLYKDYEFIY 420
421 TESKMCSTNL LKDDSSGASM DDGSPQNLPQ LRKSLHNAVR GDPTPGAGMR KTLLHHHGAV 480
481 NSRSTKYAGS DDDAGFKNSH FDEHESTERI SKLFQIHCTL EHLLMMHIHL NLANVYNDVC 540
541 SELLKKPPKL VESIDDQLSD VMDIRLSAHY VRDHLDGKDP YSRHITMRSH NKFRELKTTF 600
601 YFNSENICPP NLAQCFQCDD KEMVKERTYH SWIYHKIRSL K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
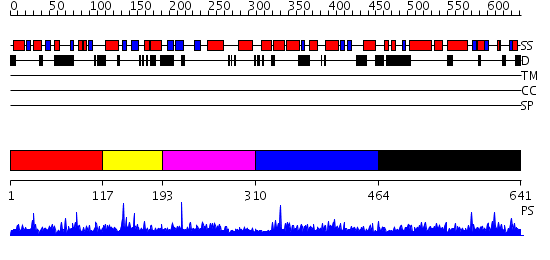
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [117..192] | 1.018988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [193..309] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [310..463] | 1.016952 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [464..641] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)