













| General Information: |
|
| Name(s) found: |
gi|24651468
[NCBI NR]
gi|23172713 [NCBI NR] |
| Description(s) found:
Found 6 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 480 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IEA]
|
| Biological Process: |
oxidation reduction
[IEA]
|
| Molecular Function: |
procollagen-proline 4-dioxygenase activity
[IEA]
oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNLERNLLQ NMRDYAEKLE EKIDLINNYV EDYNVDIEDA NEDPEKYLSN PLNSFRLIRH 60
61 MHQDWVGWQV YMEDLVAPEA VQKVESLLPQ LPQKEAFRKA ARSVHSLTQF YGYEPADLVA 120
121 KDERSSSLHL SPLDCYHLGL ELYEEQDYLG AAKWLQVAAH NYTLSRHRDL FNPLGAPRWQ 180
181 VYRDLGRTLL KLNRQCAYDA YESALRLNSQ NVHLMKEAGQ MELFSLRDPM EPILDLQPKP 240
241 TVLEKYCRGE VPRHQGRLTC ELNDWVHSFL AYAPIKFEDL QQDPFIILYP GSIYEQEIRH 300
301 VENAYERCPP NDRFELKLGI SGCSISDGYS PVLKRINERI LDMAGVEKTW DTFYIVEYAQ 360
361 LAPFEPFKLF RNSTKFPKLN LMNFEDVEAK VIIFLKDVTL GGAFTMPNGD ILVQPKRGNV 420
421 LITFENEEHS TTICPIIEGT GLGVCDSIIS HYKFKLPISL FSHDQIYYEN RRVRPPGVSI 480
481 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
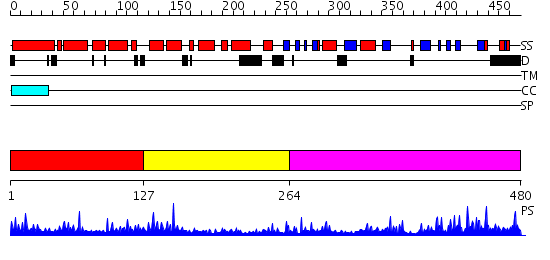
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 22.29243 | No description for PF08336.2 was found. No confident structure predictions are available. | |
| 2 | View Details | [127..263] | 14.522879 | Crystal structure of peptide-substrate-binding domain of human type I collagen prolyl 4-hydroxylase | |
| 3 | View Details | [264..480] | 5.30103 | No description for 2hbtA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)