













| General Information: |
|
| Name(s) found: |
CAP-D2-PA /
FBpp0084818
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 1380 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
condensin complex [ISS] |
| Biological Process: |
mitotic chromosome condensation
[IMP]
mitotic spindle organization [IMP] mitotic sister chromatid segregation [ISS] mitosis [ISS] |
| Molecular Function: |
binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEESHDFQFV LPLNASDLIN SSGDQYYVKE IFGAQEIPAK LQECKRKVHL GDPFYIFEHF 60
61 DLYYSIIEAR GSDGASAQNL MRSFDLLYLT VEKLFQDLQP LLTASEPMSN QQRNSYLNLT 120
121 KMTLFLQVST VKKINNSVQQ AMRDQQLNVQ KKRAKPSEGL EQFPNWEVKR GKFLVQLFNV 180
181 LQCPLEKLWS PPVAEEDFIN LLCDPCYRTI ELLPLRMDNK HVFDTIFQIL GTSIKRFNQA 240
241 MTFPVRILQI LRGTEHAAHS VAAGILLLHE EYGISSVFSI LIKSIVDALR MDSSDSSVSK 300
301 HFSNFLAEFS NIAPSLIVPH LEKLAEDLLD CQSHTLRNCV LQIIGDTVVS ELTSEDLSEE 360
361 LKEVRNEFLE HLMAHILDIS AHVRSKVLSI WHHLKTQHAI PLNFLTRVLE EAIGRLEDKS 420
421 SLVRRAAMHL IKSALESNPY SSKLSIDELR AKHEHEVQAM EKLNEVLEEE RKQEEKLNDE 480
481 FSSLAPELLP FIEENLTEFP DMQFDKEESD ETLMERIIPL MREKNYKDVI VLVRKVDFLA 540
541 GNQNMSSLLK HEEHCVYVLA LLKTYHLLAA GFKQSSEEML QQIKTVQFLK DSIDFAVLMT 600
601 SAFPKLHEML MSKTNTDVFE AVDLFTTGYM FGIHGTESGM QRMLQLVWSS DKEKRDAVSD 660
661 AYKRVLFSTD QTGRAHAIKV VQNLSKFLSE IEYGHYTALE SLMTEWVLGG DIDAAVIQVL 720
721 FERFTLKLEG TTSNESRLSL QLLIMASQSK SSIVSANTAI IEDIAVGERV RRDPRIFTSC 780
781 LQLLVNSIDA NNTAKYYKRQ NSDAEFVGKI TRLFLDFFFH RSLYDFDALA MSVFEYFYRM 840
841 CQAPDVIAQQ LVTALLKQFN ESWLVKEAAA IVPSPDKADT ETVPDSQPLE IPHSQTLTPT 900
901 QTQADSQSQM QGTLIPVYLV SRLIFCIGYM TIKEMIFLDM DIYNNMKYRD ELTALEERKN 960
961 RNQQAGSSHN AARLTLNMSA MEVRKRLSGV AAEPQQEPDD DLVGATAEDN IAEEIHGICE1020
1021 DMLLYNPDAL LSKLAPFIIE ICKRPGEFGD PTLQQAATLA LARLMTVSSR FCESNMSFLM1080
1081 NILNLTKNIR IKCNTVVGLS DLTFRFPNII EPWTGHFYAQ LHESNTELRL TAVKMLSHLI1140
1141 LNEMIRVKGQ IADMALCIVD GNEEIRNITK QFFKEIANKS NILYNVLPDI ISRLGDINLN1200
1201 LDEDKYRIIM SYILGLIQKD RQIETLVEKL CLRFPVTRVE RQWRDIAYCL GLLTYNERAV1260
1261 KKLMDNMQHY RDKVQVDEVY QSFKLIISNT NKLAKPELKA VVTEFENRLN ECLQVNPDTA1320
1321 AQGDQESTPE GQGNRTRAAP RTKKGGRKPA ANRNNTVRGR GRRARRDSTS EESFSSSSSD1380
1381 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
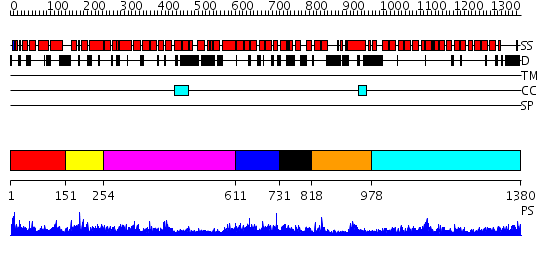
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..150] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [151..253] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [254..610] | 18.69897 | Crystal structure of Importin-beta and SREBP-2 complex | |
| 4 | View Details | [611..730] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [731..817] | 1.075995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [818..977] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [978..1380] | 88.045757 | Adaptin beta subunit N-terminal fragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)