













| General Information: |
|
| Name(s) found: |
Vha100-1-PA /
FBpp0084748
[FlyBase]
Vha100-1-PF / FBpp0084745 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 833 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V0 domain
[ISS]
synapse [IDA] |
| Biological Process: |
ATP synthesis coupled proton transport
[IEA]
|
| Molecular Function: |
calmodulin binding
[IDA]
hydrogen-exporting ATPase activity, phosphorylative mechanism [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSLFRSEEM ALCQLFLQSE AAYACVSELG ELGLVQFRDL NPDVNAFQRK FVNEVRRCDE 60
61 MERKLRYLEK EIKKDGIPML DTGESPEAPQ PREMIDLEAT FEKLENELRE VNQNAEALKR 120
121 NFLELTELKH ILRKTQVFFD EMADNQNEDE QAQLLGEEGV RASQPGQNLK LGFVAGVILR 180
181 ERLPAFERML WRACRGNVFL RQAMIETPLE DPTNGDQVHK SVFIIFFQGD QLKTRVKKIC 240
241 EGFRATLYPC PEAPADRREM AMGVMTRIED LNTVLGQTQD HRHRVLVAAA KNLKNWFVKV 300
301 RKIKAIYHTL NLFNLDVTQK CLIAECWVPL LDIETIQLAL RRGTERSGSS VPPILNRMQT 360
361 FENPPTYNRT NKFTKAFQAL IDAYGVASYR EMNPAPYTII TFPFLFAVMF GDLGHGAIMA 420
421 LFGLWMIRKE KGLAAQKTDN EIWNIFFGGR YIIFLMGVFS MYTGLIYNDI FSKSLNIFGS 480
481 HWHLSYNKST VMENKFLQLS PKGDYEGAPY PFGMDPIWQV AGANKIIFHN AYKMKISIIF 540
541 GVIHMIFGVV MSWHNHTYFR NRISLLYEFI PQLVFLLLLF FYMVLLMFIK WIKFAATNDK 600
601 PYSEACAPSI LITFIDMVLF NTPKPPPENC ETYMFMGQHF IQVLFVLVAV GCIPVMLLAK 660
661 PLLIMQARKQ ANVQPIAGAT SDAEAGGVSN SGSHGGGGGH EEEEELSEIF IHQSIHTIEY 720
721 VLGSVSHTAS YLRLWALSLA HAQLAEVLWT MVLSIGLKQE GPVGGIVLTC VFAFWAILTV 780
781 GILVLMEGLS AFLHTLRLHW VEFQSKFYKG QGYAFQPFSF DAIIENGAAA AEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
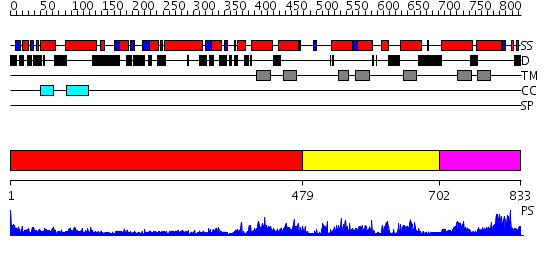
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..478] | 7.0 | COLICIN IA | |
| 2 | View Details | [479..701] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [702..833] | 3.10597 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|