













| General Information: |
|
| Name(s) found: |
ash2-PC /
FBpp0084040
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 556 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS]
|
| Biological Process: |
imaginal disc-derived wing morphogenesis
[IMP]
imaginal disc-derived wing vein specification [IGI][IMP] chromatin-mediated maintenance of transcription [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDSQMDTSS PTESSSEVNF TAEEDKSQET RSAAGVCYCG KERNLNIVEL LCATCSRWVH 60
61 ETCVSYQLGK GKLLPFITNY VFVCKNCSAS GLESFRKSQA TISQMCHCAI ANMQQAASRD 120
121 GRRQIQFSKD KEIIPYIEQY WEAMTTMPRR LTQSWYSTVQ RSLVKDVQTL FTYEEHAEHG 180
181 AMYGLFHQDL RIIKPNYESM SKSGALRLTD DGYTQASLSK NNRQKRKFPG TDSGPTGKKG 240
241 RPSSDITANV KLPPHGYPLE HPFNKDGYRY ILAEPDPHAP FRQEFDESSD WAGKPIPGWL 300
301 YRILVPHSVL LALHDRAPQL KISEDRLAVT GERGYCMVRA THSVNRGCWY FEVTIEEMPD 360
361 GAATRLGWGR EYGNLQAPLG YDKFGYSWRS RKGTKFTESH GKHYSDAYVE GDTLGFLIEL 420
421 PEEASLDYLP NTFKDRPLVK FKSHLYYEDK DKITETLKNL HILQGSRIEF FKNGQSQGVA 480
481 FEDIYAGSYF PAISIHKSAT VSVNFGPAFK YPEVLVEHKA KGMHDRVEEL ITEQCLADTL 540
541 YLTEHDGRLR LDNMGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
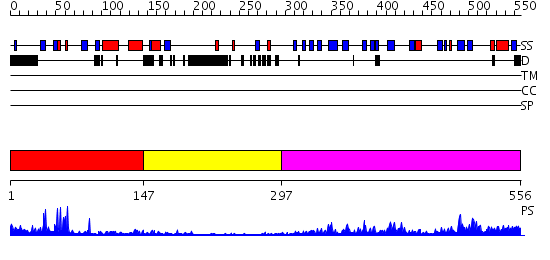
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 8.69897 | No description for 2f6jA was found. | |
| 2 | View Details | [147..296] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [297..556] | 25.0 | No description for 2fnjA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)