













| General Information: |
|
| Name(s) found: |
SMC1-PA /
FBpp0083926
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1238 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cohesin complex [IDA][ISS][NAS] chromosome [IDA] |
| Biological Process: |
sister chromatid cohesion
[ISS][NAS]
negative regulation of gene expression [IGI] |
| Molecular Function: |
DNA binding
[ISS]
ATP binding [IEA] protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEEDDDVAQ RVATAPVRKP DDETAFLEYI EMENFKSYRG HIVVGPLKQF NAVIGPNGSG 60
61 KSNFMDAISF VMGEKTSSLR VKRLNDLIHG SSIGKPVSRS CYVTAKFVLN EERHMDFQRA 120
121 VIGGSSEYRI NGESVSSSTY LNKLEKIGIN VKAKNFLVFQ GAVENIAMKT PKERTALFEE 180
181 ISGSGLLKDD YNRLKQEMIV AEEETQFTYQ KKKGIAAERK EAKHEKMEAD RYTRLQNEYN 240
241 EKQVEYQLFR LFHVERDIRK FTSDLEVRQQ EVKAVEQRKE AADEILREKK KDAGKITRDL 300
301 AKIDQEIREF ETQMNKRRPL YIKAKEKVTH CKKKLISLQK TLETAREADN AHQSDIRKLE 360
361 KQLADVEALK KRFEDEIENE SQRRGKSVNM EEGLVQEYDR LKQEAEATAT QYRSELDSVN 420
421 REQKSEQDTL DGETNRRASV EESFKKLTLQ REEAVKRRDK LMDHIKSSQA ALEEQNRIKD 480
481 ELRRDVGTSK EKIAEKQREL EDVRDQLGDA KSDKHEDARR KKKQEVVELF KKQVPGVYDR 540
541 MINMCQPTHK RYNVAVTKVL GKFMEAIIVD TEKTARHCIQ ILKEQMLEVE TFLPLDYLQV 600
601 KPLKERLRNI SDPRNVRLVF DVLKFEPQEI ERAVLFATGN ALVCETPEDA MKVAYEIDRS 660
661 RFDALALDGT FYQKSGLISG GSHDLARKAK RWDEKHMAQL KMQKERLQEE LKELVKKSRK 720
721 QSELATVESQ IKGLENRLKY SMVDLESSKK SISQYDNQLQ QVQSQLDEFG PKILEIERRM 780
781 QNREEHIQEI KENMNNVEDK VYASFCRRLG VKNIRQYEER ELVMQQERAR KRAEFEQQID 840
841 SINSQLDFEK QKDTKKNVER WERSVQDEED ALEGLKLAEA RYLKEIDEDK EKMEKFKQDK 900
901 QAKKQAVDDM EEDISKARKD VANLAKEIHN VGSHLSAVES KIEAKKNERQ NILLQAKTDC 960
961 IVVPLLRGSL DDAVRQSDPD VPSTSAAMEN IIEVDYSSLP REYTKLKDDS AFKKTHEMLQ1020
1021 KDLQSKLDVL ERIQTPNMKA LQKLDAVTEK VQSTNEEFEN ARKKAKRAKA AFERVKNERS1080
1081 SRFVACCQHI SDAIDGIYKK LARNEAAQAY IGPDNPEEPY LDGINYNCVA PGKRFQPMNN1140
1141 LSGGEKTIAA LALLFSTHSF HPAPFFVLDE IDAALDNTNI GKVASYIRDH TTNLQTIVIS1200
1201 LKEEFYGHAD ALVGITPGEG DCLVSNVYIM DLTTFEDT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
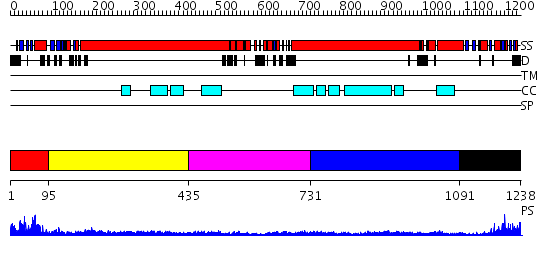
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | 23.69897 | Sc Smc1hd:Scc1-C complex, ATPgS | |
| 2 | View Details | [95..434] | 15.221849 | Heavy meromyosin subfragment | |
| 3 | View Details | [435..730] | 4.35 | Smc hinge domain | |
| 4 | View Details | [731..1090] | 2.16 | Tropomyosin | |
| 5 | View Details | [1091..1238] | 15.154902 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)