













| General Information: |
|
| Name(s) found: |
wda-PA /
FBpp0083779
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 743 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
|
| Biological Process: |
histone acetylation
[IDA]
regulation of transcription [IEA] |
| Molecular Function: |
transcription regulator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPSASNHA HAASPVYSNS NNNKSSASSK KTSSKNDLLR CAVGLLLKQK NYVSNERFRR 60
61 SDFLLLQNKQ QFAVNKMLDT DLHGGNSFTF SNVQVITNNQ HTVDQQFGRF SQFVEAQAEP 120
121 LRLEMKRFYG PMLCHFYLDL LKAREPRGAV ELLRKYAHLV APVDMYDAPP PTKINGCSTT 180
181 ANESTFNIRF AKEAQDTGDT ELDYFMRLVQ TLSGYTRLEA AESDDTVAHF RSSKYELHTT 240
241 AVVVNRICAY LQRRGHVLIM NLLYTWLHVH IVENEQRAFS EDHLLGLTDD LEGEDGEDDV 300
301 VSKPAVTLNT RGDIRPSKSL TEKSNRKRPA EEPNIMLETD IKQEVETDES AELSKLQLNI 360
361 DACLDTLKSA TEQILKSQVE LPRFLRISER SRGLTSAHLD PSECHMLAGF DNSAVQLWQL 420
421 NQSYCRGKSF YRRYPQKRCP WELNNCANQE EETDEDSSDE DVKCSEEERR ERNRARHCKY 480
481 ADNSYNEYGG FQLRGHTKGV TDVRFSAHYP LMYSVSKDAT MRCWRAHNLH CAAIYRSHNY 540
541 PIWCLDESPV GQYVVTGSKD LSARLWSLEK EHALIIYAGH TQDVECVAFH PNGNYIATGS 600
601 ADHSVRLWCA TSGKLMRVFA DCRQAVTQLA FSPDGKMLAA AGEETKVRIF DLAAGAQLAE 660
661 LKDHSASISS LSWSTHNRHL ATACSDGTLR LWDIKKLSPM SDNSSAGSSS SATTNRVLTV 720
721 NSSCQRLVDV FYGTSKTLYC IGT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
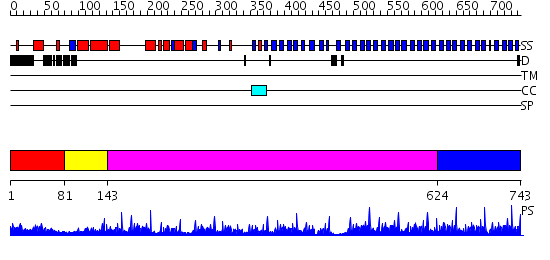
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 23.09691 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 2 | View Details | [81..142] | 14.221849 | No description for 2nxpA was found. | |
| 3 | View Details | [143..623] | 11.69897 | Tricorn protease; Tricorn protease N-terminal domain; Tricorn protease domain 2 | |
| 4 | View Details | [624..743] | 73.30103 | No description for 2ovpB was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
|||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)