













| General Information: |
|
| Name(s) found: |
loco-PB /
FBpp0083668
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1175 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
apical cortex [IDA] plasma membrane [IDA] |
| Biological Process: |
cortical actin cytoskeleton organization
[IMP]
ventral cord development [IMP] septate junction assembly [IMP] dorsal/ventral axis specification, ovarian follicular epithelium [IMP] asymmetric neuroblast division [IMP] establishment of glial blood-brain barrier [IMP] regulation of G-protein coupled receptor protein signaling pathway [IGI][NAS] glial cell differentiation [IMP] cytoplasmic transport, nurse cell to oocyte [IMP] asymmetric protein localization [IMP] |
| Molecular Function: |
G-protein alpha-subunit binding
[IPI]
GTPase activator activity [IDA] receptor signaling protein activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNNTNTTRGT CSRIPIASKQ EPTIASMVER FVQSLSQLND SGTFDSSFEI KRIHPGSSAG 60
61 STPKHVRHRC LTELDREQLT RFVQDFEERQ ASTPKVKSKI SSPYICRKAK EFYQSASGRR 120
121 SASPQITIAP ETVQEEKVLV NPLPRKCAEA EDLMPPPMAM ARVQAERRSC RSASRVLQFF 180
181 SSATKRRSGN RKLSEPEMED NRADSPVLIR VTRDRVEEQL GCSPLAASGG EVPTSSEDEE 240
241 EHDDGISSAS SNLTSQSGSS NSRNLSPDSS FEMHAPLLPS FKVTPPRAVC RVGKNAACEF 300
301 ARFLRGSFHS KRASVTTLRR SLSDPDAVQQ MDFSKPPPLR DTTNVMRNRA LPANASPFRR 360
361 AWGQSSFRTP RSDKVAKEQQ QLGQSSPVRR TASMNASDND MYIKTLMLDS DLKSSRSQHQ 420
421 LSLLQVPKVL TTPAPPSAIT ASVAAEGAAQ DHGCPSSWAG SFERMLQDAA GMQTFSEFLK 480
481 KEFSAENIYF WTACERYRLL ESEADRVAQA REIFAKHLAN NSSDPVNVDS QARSLTEEKL 540
541 ADAAPDIFAP AQKQIFSLMK FDSYQRFIRS DLYKSCVEAE QKNQPLPYSG LDLDELLKTN 600
601 FHLGAFSKLK KSASNAEDRR RKSLLPWHRK TRSKSRDRTE IMADMQHALM PAPPVPQNAP 660
661 LTSASLKLVC GQNSLSDLHS SRSSLSSFDA GTATGGQGAS TESVYSLCRV ILTDGATTIV 720
721 QTRPGETVGE LVERLLEKRN LVYPYYDIVF QGSTKSIDVQ QPSQILAGKE VVIERRVAFK 780
781 LDLPDPKVIS VKSKPKKQLH EVIRPILSKY NYKMEQVQVI MRDTQVPIDL NQPVTMADGQ 840
841 RLRIVMVNSD FQVGGGSSMP PKQSKPMKPL PQGHLDELTN KVFNELLASK ADAAASEKSR 900
901 PVDLCSMKSN EAPSETSSLF ERMRRQQRDG GNIPASKLPK LKKKSTSSSQ QSEEAATTQA 960
961 VADPKKPIIA KLKAGVKLQV TERVAEHQDE LLEGLKRAQL ARLEDQRGTE INFDLPDFLK1020
1021 NKENLSAAVS KLRKVRASLS PVSKVPATPT EIPQPAPRLS ITRSQQPVSP MKVDQEPETD1080
1081 LPAATQDQTE FAKAPPPLPP KPKVLPIKPS NWGVAQPTGN YCNKYSPSKQ VPTSPKEASK1140
1141 PGTFASKIPL DLGRKSLEEA GSRCAYLDEP SSSFV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
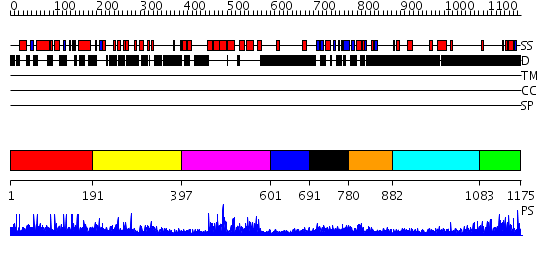
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [191..396] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [397..600] | 56.69897 | Regulator of G-protein signalling 4, RGS4 | |
| 4 | View Details | [601..690] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [691..779] | 3.7 | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | |
| 6 | View Details | [780..881] | 16.69897 | Solution structure of the Ras-binding domain of mouse RGS14 | |
| 7 | View Details | [882..1082] | 13.522879 | RBP1 | |
| 8 | View Details | [1083..1175] | 4.09691 | Fibrinogen |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)