













| General Information: |
|
| Name(s) found: |
Orc2-PA /
FBpp0082329
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear origin of replication recognition complex
[IDA][ISS][NAS]
|
| Biological Process: |
cell proliferation
[IMP]
DNA replication initiation [IDA] eggshell chorion gene amplification [IMP][TAS] mitotic chromosome condensation [IMP] DNA replication [IEA][IMP][ISS][TAS] mitotic spindle organization [IMP] DNA-dependent DNA replication [ISS][NAS] chromosome condensation [IMP] chromatin silencing [NAS] |
| Molecular Function: |
DNA binding
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASNKGGYK TPRKENLMSI ENLTNSEEES EDLNTAMVGN AVESQPKVTS RRSTRRPSPT 60
61 KKYQAYQKES NGKGQEERIV VNYVEMSDER SSDAEDQEEE ESIEESENAA RPAAKDLHLI 120
121 QSEYNVAGTS MFGFNTPKKR DAMALAALNA TPCTPKTPKT PRLGVKTPDT KRKKSMDQPK 180
181 TPAHVRTRVK KQIAKIVADS DEDFSGDESD FRPSDEESSS SSSSSDAGNS SDNDAADDEP 240
241 KTPSRARRAI VVPVLPKTPS AARLRQSARA KKSNEFVPES DGYFHSHASS KILTSDHTLD 300
301 RLKNPRLAAD RVFSLLSEIK TSAEHEGSIN AIMEEYRSYF PKWMCILNEG FNILLYGLGS 360
361 KHQLLQSFHR EVLHKQTVLV VNGFFPSLTI KDMLDSITSD ILDAGISPAN PHEAVDMIEE 420
421 EFALIPETHL FLIVHNLDGA MLRNVKAQAI LSRLARIPNI HLLASIDHIN TPLLWDQGKL 480
481 CSFNFSWWDC TTMLPYTNET AFENSLLVQN SGELALSSMR SVFSSLTTNS RGIYMLIVKY 540
541 QLKNKGNATY QGMPFRDLYS SCREAFLVSS DLALRAQLTE FLDHKLVKSK RSVDGSEQLT 600
601 IPIDGALLQQ FLEEQEKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
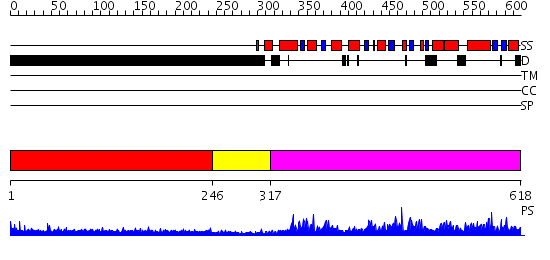
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 7.225996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [246..316] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [317..618] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)