













| General Information: |
|
| Name(s) found: |
CG3942-PA /
FBpp0082033
[FlyBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Drosophila melanogaster |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycerol metabolic process
[IEA]
intracellular signaling pathway [IEA] lipid metabolic process [IEA] |
| Molecular Function: |
phospholipase C activity
[IEA]
carbohydrate binding [IEA] glycerophosphodiester phosphodiesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHRWFFANER EECERKPEED GPSSASETQE PPPPPPVPTT EWPFCVVFHS SLNGNEYVAI 60
61 SGNCPSLGNW DPKEVYILAK NDCISCLCNC RQFEASLEIP RNIDIHYRYC VVIHDPETDE 120
121 VYIRFWESQL YPRVIRTCQN MLKNCDVFGK PHDDDEANQV DRGWATTETI VHLKIFNAPF 180
181 CWQRQKPRLL YVHVQPMFEV PENPCNEPAN PIKMVSSQTR LSRYLSTREI KAGNQYLQLA 240
241 QVEVTNLCVQ NALAAQQRFG ARCGPKDMEL FHCSIAFPEE TLYRLDLYTY AHKAGYDEPP 300
301 YHYGYGFLMP DQLLGTEGSA RVKITCASTH RPLMEMCVRY LIIRPLPNFR CDLSHSYERY 360
361 WRKNRLCMNI GHKGSGNTYR LGSDVVRENT LYGFKQAVLA NADMVEMDVQ LTQDAQVVVY 420
421 HDFVLRFMLQ RMPSFEDLLE NQDLLIFAYE NLNKLMLLAM GGSKRKDLIA VPLEAFSYDQ 480
481 LKEVKVLRFA GSKGCDKSCD RMLLEQRPFP LLLDLLDEEN LPVDMGFLIE IKWPQMTNMR 540
541 RWESGSFKPT FDRNFYVDTI LEIVLNKAGK RRIVFCSFDA DICAMVRFKQ NVYPVTLLLE 600
601 DPHSPVQYAD QRVSVQDVAV RFCNSLEFLG LTLHANSLLN KPSTMAYLHQ INLDAFVYGS 660
661 STIDLEIRNK LKKHGVLGII YDRLDQLDQV GEELEGDTMC TIDSVTTRRV IQETEVEEWI 720
721 QKCGYKPETS IVVHNIYID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
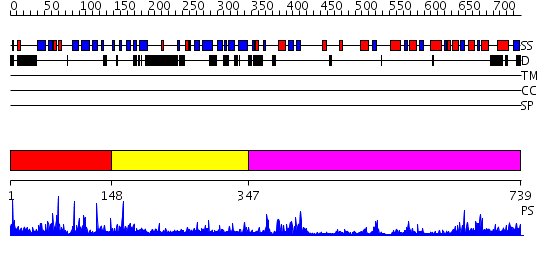
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 33.69897 | SITE DIRECTED MUTATIONS OF THE ACTIVE SITE RESIDUE TYROSINE 195 OF CYCLODEXTRIN GLYCOSYLTRANSFERASE FROM BACILLUS CIRCULANS STRAIN 251 AFFECTING ACTIVITY AND PRODUCT SPECIFICITY | |
| 2 | View Details | [148..346] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [347..739] | 50.522879 | Structural genomics, Crystal structure of Glycerophosphoryl diester phosphodiesterase from E. coli |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)