













| General Information: |
|
| Name(s) found: |
laf-PA /
FBpp0078492
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 753 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
|
| Biological Process: |
embryonic development via the syncytial blastoderm
[NAS]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRHGQHYIS ARSVSAKDLD DSESGGDKSK SKERVRNLVL KKVCRGRNVQ DSPGYRGKAM 60
61 LNCSNRTRKY SKHSKDNLHS GNEDGGMPKD TEYISSDHDD SPSWSQQSLL SSDRSKSYSQ 120
121 ICSEILEESK ERQEKAECAF RVYNINRSKL RRSHQQSLSR GPGSGSYGSS MASEYSSKSE 180
181 AGYQDYDSPS TDPSREHVQM ANVANIDKWT RPKQFKVESY KMEIKERPNY RKTQEVSHSY 240
241 SCPLESRISK GSLVTRHRAR SCVCAQESSA CSLCTAHSRS GKHHSEGCDA EVIDGPDYPV 300
301 DCRSDIPTTA TSQSRSQRDY LSSCCTRHRH HPHYHRQRSV PKTYQDRGNS PINIKTRRKT 360
361 RDYQEPHFAF RTEKVEAAVQ LSRSSASIGV QYPSQDENAD WTDWTPDTTQ GDRQFKGHDG 420
421 DCLRSTEKKR SISNEQSPIT LRNTNAKDVD IPDCFGSFAM NKHLSVITED ASQHHKDPDE 480
481 DMIDSQLSNS VLLETYDEGE KYAYSYQYSY KPEICNNNQF VSDESDLKVS SKEGYQMDQE 540
541 DYVMDKQELV HEGGSDASLS EVAKSKSFLS LKIYDADEAL MEIPEDFEGP AIVLDDDADF 600
601 LDITLTDDEE KIRAKLMAAA LTTRKTTSSI SPNISLRTRS PIEPSSLSYK PNVIFTRRSE 660
661 VIKDNYTPRP DDRVALLAEK FLQSFSESAP NDYGWKPSKQ EVTSAVSISH LFNENGVTRR 720
721 GGDTPLCGDR QLLSVEFNRK LQRQLKVIVE SFQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
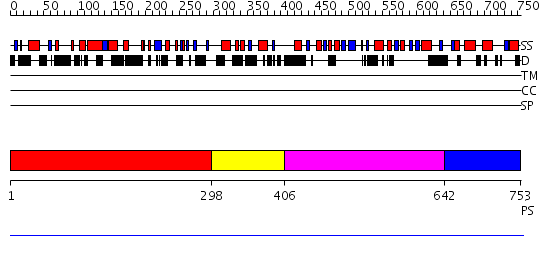
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [298..405] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [406..641] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [642..753] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)