













| General Information: |
|
| Name(s) found: |
FBpp0308243
[FlyBase]
FBpp0308242 [FlyBase] |
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 536 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
integral to membrane [IEA] |
| Biological Process: |
response to salt stress
[IMP]
regulation of transcription, DNA-dependent [IEA] regulation of triglyceride biosynthetic process [IMP] positive regulation of gene-specific transcription from RNA polymerase II promoter [IDA] mucosal immune response [IMP] stress-activated MAPK cascade [IDA] |
| Molecular Function: |
transcription factor activity
[IGI]
protein homodimerization activity [IDA][ISS] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAARLEITVS LALAATLLCS ILGHCEMSVY PGLRRRPIQP PVMDPQSEQE YAFLAQMMEQ 60
61 YARRRLQQQF DQQLHEINLK MDRQRQLLEQ ERYRQRQQEL QVQREAEEDY ESNGNNLNQQ 120
121 YDQYDNAEAE ASYGSPMEAV SDLQSPPLYL PRLPNLPPLP SLPNRPSLGN SPNRNSVLRE 180
181 RIPFAVGPSD ARFFDNPNDP SGELDSRALE DYEEYAPIEP TTEVATTASK TKIDTPVPAP 240
241 VPVEPPKLLD AANTNFHRIN PQSAAAAAVA GVNIPQSKEQ PPHELNALIN KLQKQSKAPS 300
301 HLTLVNGGDS SQEQIVLRQH LGMNSEMGVY IVALIAGVSA AVTVGLLALG VTWFHNRHKA 360
361 AADVEYPAYG VTGPNKDVSP SGDRKLAQSA QMYHYQHQKQ QIIAMENQAT DGSCGMSDVE 420
421 SDDDNEEGDY TVYECPGLAP TGEMEVKNPL FLDETPATPS NNLSSQTAGA ANAAAATNNN 480
481 ITQATPQQPA TLKKGTVKPN INLLNATGTA AAAASAASAA SATGGEEPKQ KRKSKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
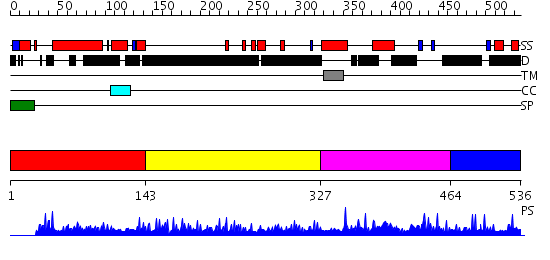
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..142] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [143..326] | 12.163996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [327..463] | 5.053972 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [464..536] | N/A | No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|