













| General Information: |
|
| Name(s) found: |
inaD-PA /
FBpp0071820
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
inaD signaling complex
[IPI][TAS]
rhabdomere [IDA] |
| Biological Process: |
protein localization
[TAS]
phototransduction [IMP][IPI][NAS] deactivation of rhodopsin mediated signaling [IMP] detection of light stimulus involved in sensory perception [IMP] |
| Molecular Function: |
calmodulin binding
[TAS]
protein binding [ISS] structural molecule activity [IDA][IMP] myosin binding [TAS] myosin III binding [IPI] receptor signaling complex scaffold activity [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQFLGKQGT AGELIHMVTL DKTGKKSFGI CIVRGEVKDS PNTKTTGIFI KGIVPDSPAH 60
61 LCGRLKVGDR ILSLNGKDVR NSTEQAVIDL IKEADFKIEL EIQTFDKSDE QQAKSDPRSN 120
121 GYMQAKNKFN QEQTTNNNAS GGQGMGQGQG QGQGMAGMNR QQSMQKRNTT FTASMRQKHS 180
181 NYADEDDEDT RDMTGRIRTE AGYEIDRASA GNCKLNKQEK DRDKEQEDEF GYTMAKINKR 240
241 YNMMKDLRRI EVQRDASKPL GLALAGHKDR QKMACFVAGV DPNGALGSVD IKPGDEIVEV 300
301 NGNVLKNRCH LNASAVFKNV DGDKLVMITS RRKPNDEGMC VKPIKKFPTA SDETKFIFDQ 360
361 FPKARTVQVR KEGFLGIMVI YGKHAEVGSG IFISDLREGS NAELAGVKVG DMLLAVNQDV 420
421 TLESNYDDAT GLLKRAEGVV TMILLTLKSE EAIKAEKAAE EKKKEEAKKE EEKPQEPATA 480
481 EIKPNKKILI ELKVEKKPMG VIVCGGKNNH VTTGCVITHV YPEGQVAADK RLKIFDHICD 540
541 INGTPIHVGS MTTLKVHQLF HTTYEKAVTL TVFRADPPEL EKFNVDLMKK AGKELGLSLS 600
601 PNEIGCTIAD LIQGQYPEID SKLQRGDIIT KFNGDALEGL PFQVCYALFK GANGKVSMEV 660
661 TRPKPTLRTE APKA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
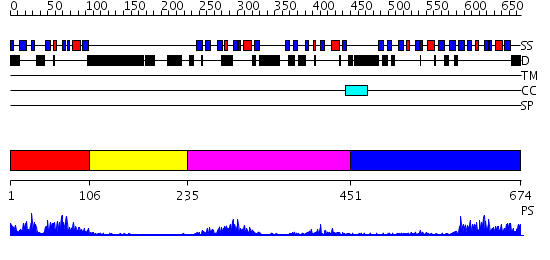
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..105] | 21.0 | Inad | |
| 2 | View Details | [106..234] | 2.34 | Solution structures of the PDZ domain of human Interleukin-16 | |
| 3 | View Details | [235..450] | 8.39794 | Protease Do (DegP, HtrA), C-terminal domains; Protease Do (DegP, HtrA), catalytic domain | |
| 4 | View Details | [451..674] | 17.69897 | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)