













| General Information: |
|
| Name(s) found: |
dnr1-PA /
FBpp0071748
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 676 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[IEA]
|
| Biological Process: |
immune response
[IMP]
negative regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria [IMP] |
| Molecular Function: |
zinc ion binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWCIVNLPNG TQQAVKWDPK ANGQECLEKV CRAMNIICEM EYFGLEHWTP NQKESQTRQW 60
61 INLRNRLSGD SGSSGSGIQL MLALRVKFWV PVHFILQESV RNLFYMQARR DLLEGRLTAP 120
121 DWSGAAKLAA LLCQADGLRF NESSLKADCP MRMRRELAQQ QQQQQQQAQQ HRLEQQRKEK 180
181 EHVLSFKKRR LSKQKSMEHI EVCTLPAAST TSCSLQPSPS ASASASASAS ASASASTSAC 240
241 SQTHSNSSSS SPSNSSSQTG LDERLASNPL RVYEEYFMQP SCEGEPPADY LRQIAVEHGK 300
301 LAKLQMSLKT AKYWLLKSIQ DLEGYGEELF SGVTTNESAT RCDIAVGAHG ITVCRGGEKQ 360
361 SIPFGAIAAA KSLRRTFKLE YVDDHNDRKE LEIKLPKQPI AAGLYRSITE RHAFYVCDKV 420
421 RGVVTNQFTR DLKGTIASMF MENTELGKRY VFDIQHTCRE VHDQARRTLH ERGGDLVAEG 480
481 AEGCSAVAGG LGASAVGEPG VSPWAMALTT GAGGSMAGKI DLAIREKEAR EAAIERCVDT 540
541 RISEAMQCKI CMDRAINTVF NPCCHVIACA QCAARCSNCP NCRVKITSVV KIYLPPELRT 600
601 SQTGSGATTT SSSSIMGDGQ VEEQLLQQQL DEISAAPASL EAGADAGVGV GGMAVGAGAG 660
661 AGATGPGGQP KVTTAA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
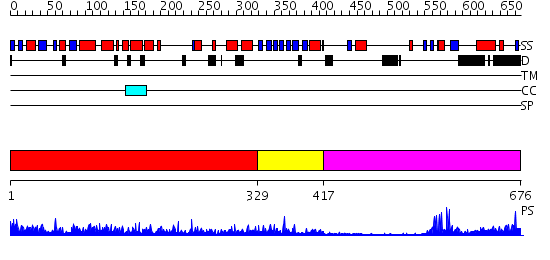
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..328] | 72.69897 | Radixin | |
| 2 | View Details | [329..416] | 2.95 | No description for 2he7A was found. | |
| 3 | View Details | [417..676] | 9.0 | Cbl; N-terminal domain of cbl (N-cbl); CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)