













| General Information: |
|
| Name(s) found: |
GM130-PB /
FBpp0071702
[FlyBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 755 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[IDA]
Golgi cis cisterna [ISS] Golgi stack [IDA] |
| Biological Process: |
Golgi organization
[IGI]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPDDTQDAKA QKLAAARKKL KEYQQRASNN NGQPGAEEPQ SALPTAASYF AQNESEHNGG 60
61 LADASSLQAI QVIIAEKAQL NAELTKVRLA CRERELELEE LRILKDQNQL RLEQLQQHCQ 120
121 DQQSTGEQQR QQYAQLQAQL LQSQAQIKDQ QSHLNELEAQ LKQSNQHQEE LQQQLSQKSN 180
181 ELEMAQLKLR QLSDESSVTT DNRVESLVQT QYMYEQQIRD LQAMVGQLTQ DKEQAAGQYQ 240
241 NYVQHQSGEI AKLNERNTEL SEVLNSLRER ERQLVEHVGA LERDIQKNIS LQAQFKEASQ 300
301 VQPSKEETSE QKALKDELAV LKEQIENFEF ERHEFQLKIK SQDDQLQLKN SALDELEKQL 360
361 ERLQAEQPDQ SKLLATMESD KVAASRALTQ NVEMKNQLDE LQQRFVQLTN DKAELVIRLD 420
421 SEEFANREIR QNYNSMEQKL QANEERFKFK DEEMIRLSHE NVELQRQQLM LQQQLDRLRH 480
481 YEAKAYHAGE ETEKENGADA PENEAEEAKE VGLLHNHSHD HSHQHSDGHC HDNDHDHSHE 540
541 HNHHNAGDHG HPHDHPHDHT HDHSHSHPHD HQHDHPHDHG QNHAHRQAAE TLPTAEAVER 600
601 LQSRFTTLIS QVADLTEEKH SLEHLVLQLQ SETETIGEYI ALYQTQRRML KQREYEKAAQ 660
661 MRLLQAEREQ LREKIEALNK LVVSLGVELP ADAAQAKQPE AAAGSEHTPG EGENSQQIIN 720
721 KIQDIISEIK ENTEQPSHNH AGDHLNCCMG KFEVV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
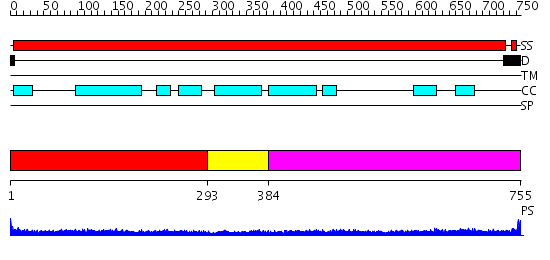
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..292] | 21.30103 | Heavy meromyosin subfragment | |
| 2 | View Details | [293..383] | 3.69897 | No description for 2iw5A was found. | |
| 3 | View Details | [384..755] | 11.30103 | Tropomyosin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)