













| General Information: |
|
| Name(s) found: |
Vrp1-PC /
FBpp0071708
[FlyBase]
Vrp1-PD / FBpp0071707 [FlyBase] Vrp1-PA / FBpp0071706 [FlyBase] Vrp1-PB / FBpp0071704 [FlyBase] |
| Description(s) found:
Found 56 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 751 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
filopodium tip [IDA] |
| Biological Process: |
regulation of cell shape
[IMP]
positive regulation of actin filament polymerization [IDA] myoblast fusion [IMP][NAS] actin filament organization [IMP] |
| Molecular Function: |
actin filament binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIPPPPGPP PPPGPPPPPA MGGLKLGGAG GADARSALLS SIQKGTKLKK TTTVDKSGPA 60
61 LSGKVCGGDG GAGIGAKRTL NNNTSTSNHN NSSSSSNGLG NGTPKLGGLF EGLSQMPKLK 120
121 PVNGIRATPS AGSAATTTSK STTNSSAQQR SNSPPAATSA SNASNGPMDF NTELTKHLTL 180
181 KRQKQQQQQQ PQPPTNNTHI NATEANLRTN RGPPPQPPKS ANSNDSILER MNIPRTAPTP 240
241 PFGNSSSVKA ASPTLSDSGS NSSGSGGGSG SVKSKAANLN ISLGNSFVNS SKSTVSASTA 300
301 SLNSSLTSST GSVRNLAPNK STLFGGSSGS SSGSSSGGGY ASVHKSSPPS TADSTPSVTP 360
361 TPPPLQLPMK PAISFGKPNF APKPPGLNQL AMANGQQRPA VTRHHSMKSP RSPPAAGIVN 420
421 GLVFPTQQHL GTLRGPLQFH GSESSSPNTS AGSMRCAPNP PKSRPSVKPP PPPTPARSFS 480
481 NTNLNNIGGS TMFSAVNTAL IQSSAISSQA PSPPITQPPS SSSSSNSVAA LRDQFRGGAG 540
541 MSNGAPSPPL PPTPAPSSNP SSASASPKSL LRDNVKPIIL NGGPLNPPAP PPHRSCPPPP 600
601 PPQRQSSNQD RVKYWIDKTV YSFKKILHIN RQGRHSGESV KVINMLNSKR NGGSGSGSAP 660
661 VPPQRHSSIR PNAPSTPPTH MANASHQFGS SSGLSSGSGA GAAASVGRLV NDLETKFGKR 720
721 FHNVTEFPKP PPFLNIQKVY PSRTFKATNG M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
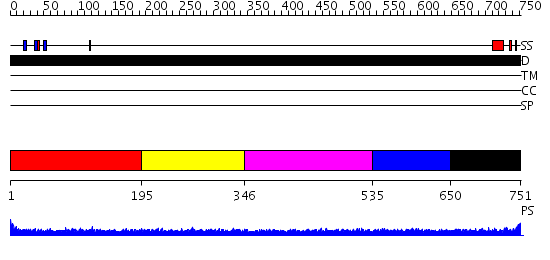
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..194] | 1.49 | CRYSTAL STRUCTURE OF THE COLLAGEN-BINDING DOMAIN OF YERSINIA ADHESIN YadA | |
| 2 | View Details | [195..345] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [346..534] | 1.154978 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [535..649] | 2.14599 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [650..751] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)