













| General Information: |
|
| Name(s) found: |
Sara-PA /
FBpp0071564
[FlyBase]
|
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1343 amino acids |
Gene Ontology: |
|
| Cellular Component: |
early endosome
[IDA]
|
| Biological Process: |
transforming growth factor beta receptor signaling pathway
[NAS]
protein localization [NAS] |
| Molecular Function: |
zinc ion binding
[IEA]
transforming growth factor beta receptor binding [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLVDIDQVL DNLELSIAAD EQLSRNAAAA AAAGANPGRF LGDGASASIS GVGAGAGVSG 60
61 AAGVTPTPAS GKSFVGVSQV FNSLDDYREN VKSLQVDAQL PSYDWQTEAE HEDDIHKFSE 120
121 QASKNNGQTI VSSSESLTTS SCSTFSSGPS PVSNGSHSEE LATPPEDEKT SEVQVEPKEQ 180
181 DQQPEQVKHE DQHQDQAVPP PTQSKQTVEE LAVGLSSISI TASGTESQSL LPEEVTASSV 240
241 LGQVLEELSE ESSPSREPET EMTNGIDVPL ANLPASVAEV PVPKEMEQPV QTEEEDVELP 300
301 QEQHPRRSQP LTFCSTMDEI SDTELDSMLQ EMDADDNQAD DQQMPDDEKS CSASPVTEEE 360
361 SVRILSDEQE TTSNDQMNVD NFSQASTVEF ADLKQPESTP VTAMGEDVAS SFSSTESDSL 420
421 SGRKLDGEVE VDREHEDASL ATDALETQEQ RPQRPQTLNL NGCQAESSTP SQDENPEIQQ 480
481 ETLQPTEVRE GAAGVGVIPG EDDEESPIYE AVGYSDPHAN LGKVPPIWVP DNMAGQCMQC 540
541 QQKFTMIKRR HHCRACGKVL CSVCCSQRFR LEFATEPESR VCVQCYMILS ERQANGLNSE 600
601 SAPGSALPPT PIRSPNPNNP MEYCSTIPPH RQVANSPGAP PPSVIVPVGV LKKTDGSSSN 660
661 SSSSDGQKGQ RKRKSVMFSD GIAPGSELAS TMEQQWGESK HSRRGLSRSG SGAGQKPPTP 720
721 ATEEPPRSID TSSTLGLVAQ LFRGSIPPSA AAATLSATET SAGRSSSQSQ AQTSPRRKAE 780
781 SSRNRKLPPN GDAQGCYLPP EENGLPPICI SSKEGGEVDY KVVRNDGSLL DRLQNETLKF 840
841 IIKNNFFVLV KIVNMSCCMN RWVLNFTTSG LHHMGSDELI ILLEIKQPKQ EEYVKGEVAI 900
901 PKDLFQHLYE IYVEAEHARS GTHELAFTSP RNANFLGSRE HGGFIFIRPT YQCLQGVIVP 960
961 DNPYLVGVLI HRHEVPWAKV LPLRLILRLG AQYRYYPCPL ISVRGRESVY GEIAQTIINF1020
1021 LVDFRNYSYS LPAIRGLYIH MEDRQTTVII PKYRQQDVIK AINNASDHIL AFAGNFSKVA1080
1081 DGHLVCMQNI NDDRSEMYSY STQAINIQGQ PRKITGASFF VLNEALKSSS GLSGKCSIVE1140
1141 DGLMVQIMPA KMEEVRQALR NQNDIDIVCG PIDATDDQSE IVSIKWVDNN RDINVGVKSP1200
1201 IDDQPMDGIS NMRVLKWASF NYSNTNYAIR LSDIYIIKCE DWYATNGGNC EEITRIAEQL1260
1261 ARSASMALLP LLDLLAAAGI NKLGLRATLD QDNVGYEAGA RGSKLPPLYM NALDNHLIAT1320
1321 LHGESSGIQE PIILELIFYI LNA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
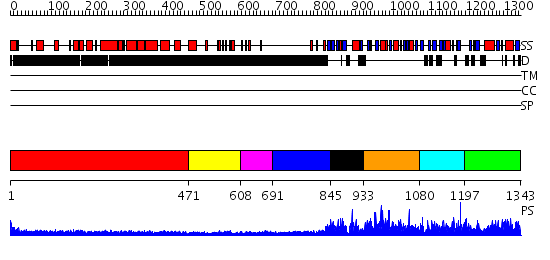
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..470] | 4.045757 | COLICIN IA | |
| 2 | View Details | [471..607] | 3.54 | Hrs | |
| 3 | View Details | [608..690] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [691..844] | 3.522879 | Structure of the full-length E. coli ParC subunit | |
| 5 | View Details | [845..932] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [933..1079] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1080..1196] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1197..1343] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.77 |
Source: Reynolds et al. (2008)