













| General Information: |
|
| Name(s) found: |
FBpp0309676
[FlyBase]
FBpp0309675 [FlyBase] Magi-PA / FBpp0071559 [FlyBase] |
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1202 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[ISS][NAS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein binding
[IEA]
guanylate kinase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPIITATTTS TAGGGAGVAA RGVATAKSPT DYLMLQQQQQ QQQQQQQHFN SNHFYQQQQP 60
61 QQLAPLNASH QHQHSHPHPH PHPVAAATSF ATFNGNSNLN SSLANGSSRK FNCSRDNVDD 120
121 GGDAGEMAAA SQTLSSAGTD VVGNGSGHGN GTVTGNGEKS GGLAPPGGHH LRPPPPPALK 180
181 KSASAVIGNS ASREDVASLS GTSSLNSQMS VHNQFISGLP SNGIGSNGVH GGAATGRGGP 240
241 PNSILKGSKE NLLQNTYYNG ETGDSSLGSE CGVGDVQHQN HESHDDPGDG LGPLPPKWET 300
301 AYTERGELYF IDHNTGTSHW LDPRLSKYQK KSLEDCCEDE LPYGWEKIED SMYGMYFIDH 360
361 VNRRTQYENP VLEAKRRAAE QSQQQQQMQQ QQQQMQEQRS KTPTALSETM PTEAAEAHEQ 420
421 EEDESPMKLP YKFTRNPAEL QGQRINTTLL KSSRGLGFTI VGSDGSAGGD VEEFLQIKTV 480
481 VPNGPAWLDG QLQTGDVLVY VNDTCVLGYT HHDMVNIFQS ILPGERAALE VCRGYPLPFD 540
541 PNDPNTEVVT TMAVDGRESD KQRRLNMDGN YNFLDLSGEG AKKTSGSGSG FILMKKPEIY 600
601 TFSIMKGSMG FGFTIADSAC GQIVKKILDR NCCTQLMEGD VLLEINGLNV RNKPHFYVVE 660
661 LLKECSQTTP TAVKIQRTPP DPPANNTLAQ LNQVGNVAKL RKNFVGSGLF RSKTPTADLY 720
721 STQVKEVLPM RPKTPLVDTR RSRVQIQSPN NEVDDEGDGA AAAERKPLQL QTQGKSNSSL 780
781 QELDDIPYMD PYPKISRLSE RLAEVTLQGD ANGGIYGMPP SMQPLPLPLA HHESCYCYDC 840
841 QAQRYRPGYF VQAQQAAMSG TTGQYSPLQT AHLQNERIQR RVNELLSDRR RVGFANLDPP 900
901 QQMQHSPSWR NGALLDVSED ADQCELTEVT LERQALGFGF RIVGGTEEGS QVTVGHIVPG 960
961 GAADQDQRIN TGDEILSIDG INVLNSSHHK VVSLVGESAL RGQVTMILRR RRTPLLQQAP1020
1021 VSTQLRRYPY DVIVSRHENE GFGFVIISSS NHYYGSTIGK LIPGSPADRC GELKVGDRIV1080
1081 AVNRIEIAGM SHGDVVNLIK ESGLHVRLTI GVPLKEGGPS PGGSSVGVSS NTPLQASPSL1140
1141 LKAQLSHQQQ LPQQHLQQAQ LNHHHQHLEM PMSMPVPGHG SYLERPSNNI SATLALHQQP1200
1201 QL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
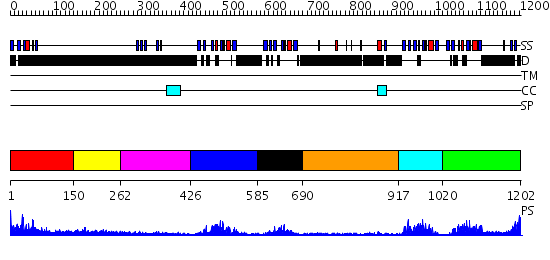
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 27.221849 | No description for 2nq3A was found. | |
| 2 | View Details | [150..261] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [262..425] | 4.48 | NMR structure of WW domains (WW3-4) from Suppressor of Deltex | |
| 4 | View Details | [426..584] | 18.522879 | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) | |
| 5 | View Details | [585..689] | 10.154902 | Solution structure of the second PDZ domain of human membrane associated guanylate kinase inverted-2 (MAGI-2) | |
| 6 | View Details | [690..916] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [917..1019] | 4.02 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 8 | View Details | [1020..1202] | 12.0 | Solution Structure of The forth PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 |
|
|||||||||||||||
| 4 |
|
|||||||||||||||
| 5 |
|
|||||||||||||||
| 6 | No functions predicted. | |||||||||||||||
| 7 | No functions predicted. | |||||||||||||||
| 8 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|