













| General Information: |
|
| Name(s) found: |
CG4966-PA /
FBpp0086017
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 858 amino acids |
Gene Ontology: |
|
| Cellular Component: |
late endosome membrane
[IDA]
lysosomal membrane [IDA] |
| Biological Process: |
dTMP biosynthetic process
[IEA]
miRNA loading onto RISC involved in gene silencing by miRNA [IMP] negative regulation of gene silencing by RNA [IMP] transport [IEA] siRNA loading onto RISC involved in chromatin silencing by small RNA [IMP] |
| Molecular Function: |
odorant binding
[IEA]
FAD binding [IEA] thymidylate synthase (FAD) activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRDWRKTKH KRWQKVQQKQ QTSILTERET MIVFVYDTEC LTDEADDPIS AVLYFHPSWV 60
61 SDSQKVALCG QLMGTSYFLK DCFFNPRILA LQNGKFVLKE FGRFILAIGT DRNIGDQLLE 120
121 HRADLLSSLL KFFHRDVQTL YAQYAAPPAL NRRNLSEKLY HIFETYLPML QRNGNTFQNV 180
181 PRLRMPKTAS HIFLEAIQTL QSCQQTKGIL GGAILYHNKV VASQLSDMVT KHLVLTDPLH 240
241 IRTAAEQVTN HPEFHIPNGV QMLVVYLEHM QYRQLAGEAQ RAQNLQMNTA QLTQNGMPFQ 300
301 YAKRKIKRDK SLIFTHIPEE EHAPEQQGAV EQLPPARPKS MRPTHLPLRI KSMQSKELPE 360
361 SGIASINFDE TDSYPQFIGR TSVCNTPMTE NKVLPVANVM SICANPEDEG KEEDIHNSNG 420
421 KTHSRRNSLK VDVEKFFQNF ISNPNKQLTR RKSSADLQDA LRAISKKLNN FTHGLKTDVN 480
481 RNGSGNGDVS SDSPDFIEDD DKITSRTISD PTYPVFNTNG QQISRSLFQQ FLDQYRKLWG 540
541 VASEQAHEDA ELAALVAEFQ EFNAEIQKLD EHMRQQAAEA SSADRNLNVS AAKTPLDKRS 600
601 MTLPLKSAGE STFGERASGR SGAGGVPLTP LMAKLSVLAL SETTPIEIQT PLTTSKVFPR 660
661 RSSLKCEDAV DALAALTTAP AQPPGPIQPD GLQRTELYIC GQQNMTLLLL MEEGTCQQQQ 720
721 VVQKMFDICV AKFPHMESQL NQTLNVNVEG DNRDGSNYSF MCVDSKWDVL QRNGPWNPLE 780
781 LNILESMHSI HSSGHHLTDL ILRSNDSVYY GHKNGRTEFF YKEPTHQING IPPPSDPIGN 840
841 IQSRAKSRLE RDHSYMLF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
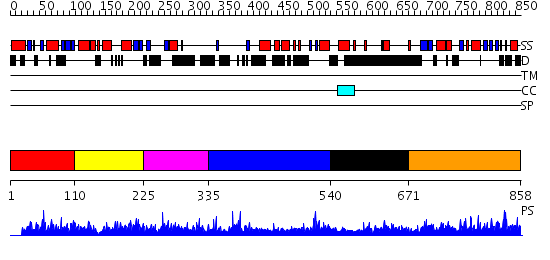
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..109] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [110..224] | 1.014973 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [225..334] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [335..539] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [540..670] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [671..858] | 2.011933 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)