













| General Information: |
|
| Name(s) found: |
gi|28573498
[NCBI NR]
gi|28380767 [NCBI NR] gi|21429090 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 825 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[ISS]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHQTEDLKLI DRQLSKIARK RYKWQEKNKT TFVLCTHGMV NDDDKVKQCN PEEKQQQLLS 60
61 MMRELPYYPP SNEMIEANTN QDLSANNSVF GDFMKTKRLE FYNVCSVMQT LLTIDDLSNM 120
121 ELYAQLIQSD VPVQRVSKLT YKITFKSTPV NAEDFVAPNL DQVVLVPKAS LLASPLPKQL 180
181 VLALLPHPHE DLEPGDPMLR HVASVDKVSS TTVHVRFSRK HKAVLSGQRF YAIIRSRRLT 240
241 IRYMYRALNL LQESPLVRRY LFPFPSWLTQ LVDNSQIKVC CDGHLPNWMQ GRPQEPATPT 300
301 SLSLLNSTIY SNAEQLEAVQ RIVAGPSTQG PYILFGPPGT GKTTTIVEAI LQLRLQQPQS 360
361 RILVTAGSNS ACDTIALKLC EYIESNIRLQ EHFAQQKLPE PDHQLIRVYS RSIYEKGFAS 420
421 VPSLLLKNSN CSKSIYDHIK ASRIVKYGII VATLCTVARL VTDTLGRYNF FTHIFIDEAG 480
481 ASTEPEALIG IMGIKQTADC HVILSGDHKQ LGAVIKSNRA ASLGLSRSLM ERLLQSDCYK 540
541 SDENGNYDRN RQMRLCRNYR SHPQIVRLFN ELYYNGELKA QAPAMDVNLA ANWSVLTNPQ 600
601 FPIIFQATHG VTNREQNSTS SYNNLEAEVI CWYVKRLIND RVVGQEDVGI VAPYTAQGKL 660
661 VTKLLQSKGY PNVEVGSVET YQGREKTIII ASLVKSFTNM GFMCNPRRVN VLLSRAKALL 720
721 ILVGNPVTLR HHSDFKFVIN ECKKHGTYLL KKRDSGQRPH ILIKADESNE ESSSEPEDED 780
781 DKLIPSYARM QQHTDAAIHK IKAHPTNEND QLANSFKISM SKLSI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
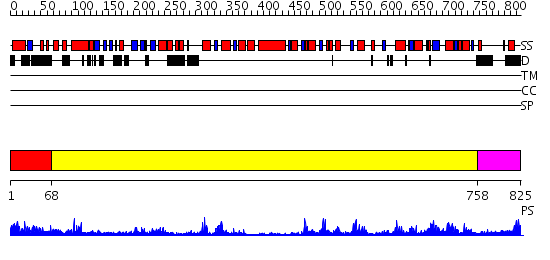
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..67] | 17.522879 | No description for 2iykA was found. | |
| 2 | View Details | [68..757] | 97.522879 | No description for 2gjkA was found. | |
| 3 | View Details | [758..825] | 81.522879 | No description for 2is1A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)