













| General Information: |
|
| Name(s) found: |
Syn2-PC /
FBpp0086242
[FlyBase]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
syntrophin complex
[ISS][NAS]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
structural constituent of muscle
[ISS]
protein binding [IEA] cytoskeletal protein binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLSDTMATP IEEKLDQNLK VRTGMVHVSD GKSRPEPLRL NLTMELLSLQ KLEVVTPSSC 60
61 HPNGPPMESK ERMVQITRQK QGGLGLSIKG GAEHKLPILI SRIYKDQAAD ITGQLFVGDA 120
121 IIKVNGEYIT ACPHDDAVNI LRNAGDIVVL TVKHYRAATP FLQKQLPLMM AYVTRYIYGT 180
181 DKLRQNAFEV RGLNGISTGV IHCDELAILS QWLKLITDNV VGLTHLQMKL YNRNFAVGER 240
241 IEYMGWVNEG VINNNISWQS YKPRFLLLKG TEVMLFETPP LNVAGISKAL VVYKVYQTMF 300
301 RIVKESETVD SRQHCFLLQS SGHEPRYLSV ETRQELLRIE NSWNAAIVTS VIKLGRKTFA 360
361 VSHHGKSGGL TLDWQAGFSL TEGAESATVW QYKFSQLRGS SDDGKSKLKL HFQDHDSRAI 420
421 ETKELECQVL QSLLFCMHAF LTAKVASVDP AFLSSIQQT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
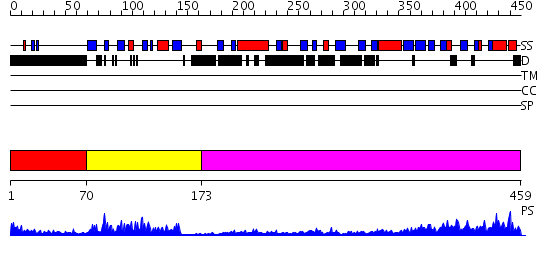
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | 26.09691 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 2 | View Details | [70..172] | 12.30103 | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | |
| 3 | View Details | [173..459] | 16.0 | Psd-95; Guanylate kinase-like domain of Psd-95 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)