













| General Information: |
|
| Name(s) found: |
Pms2-PA /
FBpp0086520
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 899 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
mismatch repair
[ISS]
|
| Molecular Function: |
ATP binding
[IEA]
mismatched DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLETNYNADT EEEPDVPPPT TALSGQIKAI GKDTVHKICS GQVVLSLAVA VKELVENSID 60
61 AGATLVEIKL KDQGLQSVEV SDNGSGVEEM NLEGMTAKYH TSKIREFVDL LGVETFGFRG 120
121 EALSSLCALS DMVIQTRHKS TDVGVKVELD HEGRIKKRSP CARGVGTTVL LANLFSTLPV 180
181 RRRDFTRNIK KEFTKMCQIL QAYCLVTKGV RILCSNHTPK GAKTIVLQTH GDQEVMANIS 240
241 AIFGARQAAD LVPLKSPFGQ GQLTEAELRA DLESGADLAD TTSPQISTED VERLNQADFQ 300
301 LEGFISSCRH GAGRSSRDRQ FFFVNSRPCD PKNIAKVMNE VYHRYNVQQQ PFIYLNIITA 360
361 RSDVDVNLTP DKRQLLINNE RILLLALKKS LLDTFGQTPA TFQMQNTTIV SMLEPKTNSG 420
421 KTKFPKESSK DDQHEEVSEE DVPTTSTQRF MDVLTQWRRT GDTKGTAPSV PVKRRCSESE 480
481 ELVTRTLKMQ KIHTFLSQES PNEQSSKCEA ESDAASDVDM AKKDKTQVSL DNSFLNLKEL 540
541 AKESEAYDLL TKPQNTPRID CKVLTPIKSR RSIFEFKTDA MALDKQESPA ELNSQPPSFT 600
601 QLSEETDIAS DEDDHIELPT RIEFDEEMEE GLPSNFSSGE ITTSLEEIAS SLKAHEQQQR 660
661 DRRTRTKLQR LRFKTEINPN QNTSAEAELQ REIDKEDFAR MEIIGQFNLG FIIVKLEDDL 720
721 FIVDQHATDE KYNFETLQRT TQLEYQRLAV PQNLELTAVN EMVLLNHIDV FEKNGFKFEV 780
781 DHEAPATKKV RLLGKPHSKR WEFGKEDIDE LIFMLQDAPE GTICRPSRVR AMFASRACRK 840
841 SVMIGTALSR NTTMRRLITQ MGEIEQPWNC PHGRPTMRHL INIAMLINSD ENDEQEPPA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
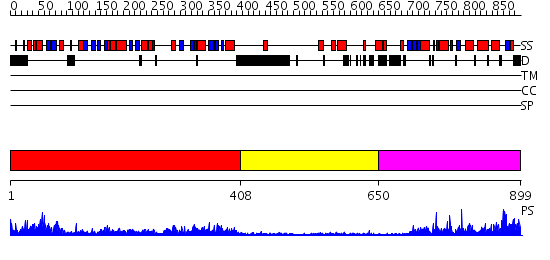
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..407] | 73.154902 | DNA mismatch repair protein PMS2 | |
| 2 | View Details | [408..649] | 6.69897 | No description for 2cgeA was found. | |
| 3 | View Details | [650..899] | 24.69897 | Crystal structure of the MutL C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)