













| General Information: |
|
| Name(s) found: |
CG8485-PC /
FBpp0086698
[FlyBase]
CG8485-PB / FBpp0086697 [FlyBase] CG8485-PA / FBpp0086695 [FlyBase] |
| Description(s) found:
Found 51 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 860 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein amino acid phosphorylation
[IEA][NAS]
|
| Molecular Function: |
ATP binding
[IEA]
SAP kinase activity [ISS] protein serine/threonine kinase activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLETQPVGG VSDGKIAGLY DLEETLGSGH FAVVKLARHV FTGAKVAVKV VDKTKLDDVS 60
61 KAHLFQEVRC MKLVQHPNVV RLYEVIDTQT KLYLVLELGD GGDLYDYIMK HDSGLSEELA 120
121 RKYFRQILRA ITYCHQLHVV HRDLKPENVV FFEKLGLVKL TDFGFSNKFL PGQKLETFCG 180
181 SLAYSAPEIL LGDSYDAPAV DIWSLGVILY MLVCGQAPFE KANDSETLTM IMDCKYTVPS 240
241 HVSTDCRDLI ASMLVRDPKK RATVEEIASS AWLKPIDEPD STTSTSEHFL PLVSREQLGE 300
301 EDHAFIIQKM INGNIASKEE ILQALDKNKY NHITATYFLL AELRLRRRRE ELAQKQRLLN 360
361 DASMKVGDAS RKLVPEKPSP TEAQKGGVPI SINVTPAAQF ANDGGKPDKR SRKCSIVREE 420
421 DEEESATECT AVGNELKVTS SSRREAFSDS PFSRSIHERS SSEPGNQTKG EQIANSGVPS 480
481 HSRTKIVVSV DATLAHKLKQ MEKSAKIDED TLSGLKELEI GKLKPLPSSS KNPVLTHRRT 540
541 KLNKIRTPSC SSSEASDDDT KTRNKKKINK FVGDTPVRFR MHRRDSHDDS SDSQDQLYPP 600
601 PGSASNAGNF ISKSGSVSGK KEGQSQYKEP NNKSDENRKS HSQKNRKQHN KDHVDKHSHA 660
661 ERQLADSSTA TTGNSTNRRR RIRESQSLDR ITEAQEYEMR HRCYAESEAA SSSSSSTQHI 720
721 DQRYSLLSLN TSTFSVAETK EEYDEEADAT NATDQIMNTL NAAKATLQQE QYFNNNTNLS 780
781 RNFNHNHNQS IIHISSQKAN GKKSNETPKD IYNLNSIEHI DLILEDKSLT KAIHVTGSKC 840
841 FVTMKKIRRL GKYFPVMNFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
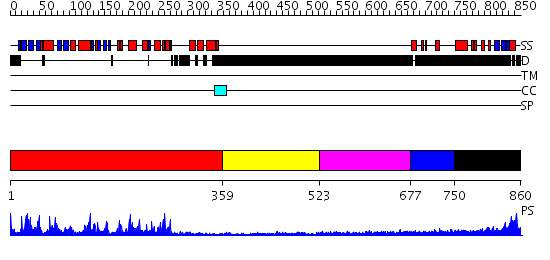
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..358] | 93.09691 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 2 | View Details | [359..522] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [523..676] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [677..749] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [750..860] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)