













| General Information: |
|
| Name(s) found: |
dgo-PB /
FBpp0087345
[FlyBase]
dgo-PA / FBpp0087344 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 927 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS]
|
| Biological Process: |
establishment of planar polarity
[IMP][TAS]
establishment of imaginal disc-derived wing hair orientation [IMP][TAS] establishment of ommatidial planar polarity [IMP][TAS] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQHGSSRIPK QEALPNAISG PDSSNWTSLH EAVAAGDDRR VEGLLFSNAD RLARESVQGN 60
61 TPLHEAASRG FSRCVKLICT PPTPPASSGV GKAAKGQKDK DKDKNKGRVK VAHQTIEALH 120
121 NSALGIANNE GLSALHLAAQ NGHNQSSREL LLAGADPDVR NNYGDTPLHT ACRYGHAGVS 180
181 RILLSALCDP NKTNLNGDTA LHITCAMGRR KLTRILLEAD ARLGIKNAQG DCPMHIAIRK 240
241 NYREIIEILN TPKKIRNRKE KPKEGGSRSD KDRDRERDVV DKGINWSPYG CHYFPDPRAF 300
301 PSPKLETLPK EPLKSGEQYF LDLAGHIHKG PVSVGNTCYC GPFFRHIENK LNCNRKSLKK 360
361 YVHKTKERLG HKVQALAIKT NDQIEQLTRT MIEDRLRCES KRQHLSEFLR RGEPMRSTFD 420
421 QQNKSSRIER TMSRCRSLEL LENNHEGGRL TNSRSVDVLE DQAVEAIVHR SAEVQVDSDS 480
481 DDDSNEARDE EEAEVGDKDQ VQEEAEPEEH SKLDELKLDF LKVSERLGVL LEKTTLIMER 540
541 DSEQESKRQL SSLSPPYNPH PRAESAQLRE DKESVNSPNF DEYANVIRRY PNSSETSNPS 600
601 QNSNSWDWEA SPTGSNPHPD YNKYYQRIGR GENMLNSVIK ALRKDASFAD LGKEETAVNE 660
661 TAEGMGGDKL LYKEQASEAA GISNLMKRQP LCLEDNYPVM SNLFYSNPIM EYAEETEVRQ 720
721 NGNESMSKVE IRNSEIKCLK KPNAGQVRDM VAQLQNTIDS NRQDSQTAHV IAARSHSRLN 780
781 NNFHHLEGTS SNPSPTYSIP HRSSAQFGLP ERHIPKDAYF HELPHRPPIP QTPQRRLHSG 840
841 YVPNASLYRS EDLYVGRIPH PNAPPRQVDY VDAVAYRPLQ SPTFQAIVPP HVNQRPRNWP 900
901 HPNLPSNEID LDELSAVGLY NNVSSLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
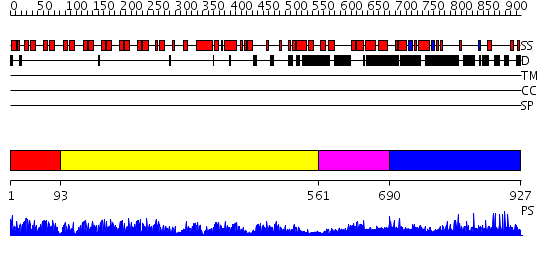
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] | 56.0 | Crystal structure of ribonuclease | |
| 2 | View Details | [93..560] | 103.0 | Ankyrin-R | |
| 3 | View Details | [561..689] | 1.169985 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [690..927] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)