













| General Information: |
|
| Name(s) found: |
pgant3-PA /
FBpp0088130
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Golgi stack
[NAS]
|
| Biological Process: |
regulation of cell adhesion mediated by integrin
[IMP]
protein amino acid O-linked glycosylation [IDA][IMP] oligosaccharide biosynthetic process [IDA] O-glycan processing [IDA] |
| Molecular Function: |
acetylgalactosaminyltransferase activity
[IDA]
polypeptide N-acetylgalactosaminyltransferase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGLRFQQLKK LWLLYLFLLF FAFFMFAISI NLYVASIQGG DAEMRHPKPP PKRRSLWPHK 60
61 NIVAHYIGKG DIFGNMTADD YNINLFQPIN GEGADGRPVV VPPRDRFRMQ RFFRLNSFNL 120
121 LASDRIPLNR TLKDYRTPEC RDKKYASGLP STSVIIVFHN EAWSVLLRTI TSVINRSPRH 180
181 LLKEIILVDD ASDRSYLKRQ LESYVKVLAV PTRIFRMKKR SGLVPARLLG AENARGDVLT 240
241 FLDAHCECSR GWLEPLLSRI KESRKVVICP VIDIISDDNF SYTKTFENHW GAFNWQLSFR 300
301 WFSSDRKRQT AGNSSKDSTD PIATPGMAGG LFAIDRKYFY EMGSYDSNMR VWGGENVEMS 360
361 FRIWQCGGRV EISPCSHVGH VFRSSTPYTF PGGMSEVLTD NLARAATVWM DDWQYFIMLY 420
421 TSGLTLGAKD KVNVTERVAL RERLQCKPFS WYLENIWPEH FFPAPDRFFG KIIWLDGETE 480
481 CAQAYSKHMK NLPGRALSRE WKRAFEEIDS KAEELMALID LERDKCLRPL KEDVPRSSLS 540
541 AVTVGDCTSH AQSMDMFVIT PKGQIMTNDN VCLTYRQQKL GVIKMLKNRN ATTSNVMLAQ 600
601 CASDSSQLWT YDMDTQQISH RDTKLCLTLK AATNSRLQKV EKVVLSMECD FKDITQKWGF 660
661 IPLPWRM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
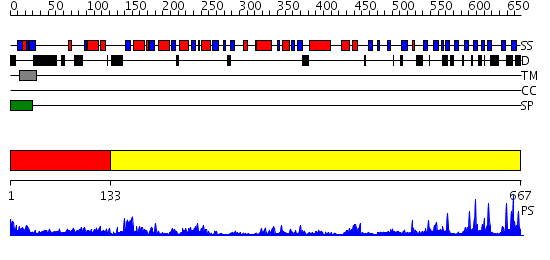
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [133..667] | 95.522879 | The Crystal Structure of UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase-T1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|