













| General Information: |
|
| Name(s) found: |
Tdc2-PA /
FBpp0085475
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP]
behavioral response to cocaine [IMP] cellular amino acid and derivative metabolic process [IEA] oviposition [IMP] carboxylic acid metabolic process [IEA] |
| Molecular Function: |
pyridoxal phosphate binding
[IEA]
tyrosine decarboxylase activity [ISS] aromatic-L-amino-acid decarboxylase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSTEFRKRG MEMVEYICNY LETLNERRVT PSVEPGYLRH LLPPEAPQEP EDWDQIMRDV 60
61 EDKIMPGVTH WQHPRFHAYF PAGNSFPSIL GDMLGDGIGC IGFSWAASPA CTELETIVLD 120
121 WLGKAIGLPD HFLALKEGST GGGVIQTSAS ECVLVTMLAA RAQALKRLKA QHPFVEEGHL 180
181 LSKLMAYCSK EAHSCVEKAA MICFVKLRIL EPDDDASLRG QTIYEAMEED ELQGLVPFFV 240
241 STTLGTTGSC AFDNLPEIGK QLQRFPGVWL HVDAAYAGNS FICPELKPLL KGIEYADSFN 300
301 TNPNKWLLTN FDCSTLWVRD RIRLTSALVV DPLYLKHGYS DAAIDYRHWG VPLSRRFRSL 360
361 KLWFVLRSYG ISGLQHYIRH HIKLAKRFEE LVLKDKRFEI CNQVKLGLVC FRLKGSDKLN 420
421 EKLLSIINES GKLHMVPASV GDRYIIRFCA VAQNATAEDI DYAWDIIVDF ANELLEKEQH 480
481 DELSEIMNRK KQDTLAQKRS FFVRMVSDPK IYNPAINKAG TPKLSMELPS PVVSRGSAPI 540
541 IRTQSSVDHN SWISWPLAFL FNSNNEEKGS NVSLRFRHLD TNVRPSSSRR NSGAGSSPSP 600
601 ENELDYVNVQ QQQMEQRSPR RSPMAVRKAS STRDNLN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
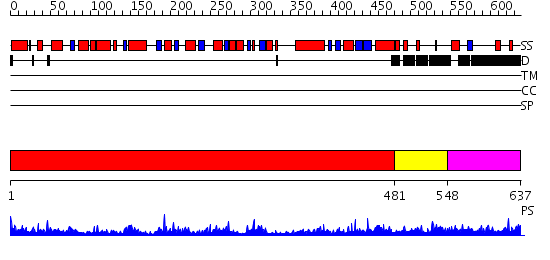
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..480] | 115.0 | DOPA decarboxylase | |
| 2 | View Details | [481..547] | 30.69897 | Human mitochondrial serine hydroxymethyltransferase 2 | |
| 3 | View Details | [548..637] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)