













| General Information: |
|
| Name(s) found: |
jing-PD /
FBpp0290850
[FlyBase]
jing-PH / FBpp0112913 [FlyBase] |
| Description(s) found:
Found 42 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1486 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS][NAS]
|
| Biological Process: |
central nervous system development
[IMP]
specification of segmental identity, maxillary segment [IEP][IGI] axon guidance [IMP] border follicle cell migration [IMP] imaginal disc-derived wing vein specification [IMP] regulation of transcription [NAS] tissue regeneration [IMP] open tracheal system development [IMP] leg disc proximal/distal pattern formation [IMP] glial cell migration [IMP] |
| Molecular Function: |
transcription factor activity
[NAS]
zinc ion binding [IEA] transcription repressor activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPPHILGSAS GSSSTAAASP PTSSASQTPP SPAIHHSHLS QHNATSAMSK PTTPQRHGNP 60
61 SLLPTLQWPW NTSLGSTSTA VPAASANKNK RTAAGSGAAL GLSGEGSCLT GGGAASAKKA 120
121 RSDLPGSFDA SKRLKVAAME ESQTKITGFF KSQMKPSPGG GKLSPQPGQQ SNPANTLTMS 180
181 TPATTASLNK YFNILSQLKE QKAQQQQQQQ QASKTLPTPA PVAPVPSAPP VAPVVVTPSP 240
241 SLPVPALKKI ERSNKQPAKI AQVAPNLRKT PSSGSSGSSS SSSSSSSNGS NTGKSPTKKH 300
301 VAIAPRTPEM KQQQQQGKAA MVYRPPVTST ALKQKQISPP APVAAHTPAP APALNPTPAP 360
361 ANPTLYQLPV QLPNLVQLPP QLAAAANIMQ LNNVAKAAVA AAATNNAAAQ AAQAAQAAAA 420
421 QYFLNGTVFK LQQVTTATTT TATTATAAAA PAGNPFGLLN LATAGNPFGL PNANGQFPIE 480
481 AIPGAGSYLH HQLLLARQNN MSLNETMASN SCGNMNFVNP LNNQQVGLKD NKLYIVNSAE 540
541 YLGYLMSLQI ALNNQQHQQQ QIISATQPPP LAATNNNSTN TATQPPPLAA TNNTNSNTAN 600
601 NSTASNSINH NASNNLSNIN NTAIQPQRAI VKPPLHSSTQ PPPLVTISSM PACSPAAASS 660
661 PAAKRSLPAA KPYQKRLTAK GRVGTAPIIA TPTTPPPLVP TSATKELGQL RKSTGTTGTP 720
721 TGTPTPTPPL VSIAPSKLTP TLSVSKQGPT MKLANSAPDL FDLVKNSKLV AKVSQPLTPL 780
781 PFSSPSSSSN GSHGSHGMRS SPALSTSNSC TLSAFSKIKV ETTELASQTG SLTSSSIPTI 840
841 SLKPQSFAGQ LPKREPESET DTLKHDLLPD CTNSNSNSNS CSSSTYSHSV SSAADLSLEA 900
901 STPAPSPSPS ASPSGLGSPS PAASNLSASS RRAASQTDML SELVTSSCIS SGGDDCSQAT 960
961 DSPPMPALPL AKSEDATTPI STVSGGSSSG SSNYDEEDDK SVASLETHQT HKRLRDLPTP1020
1021 ESGIGGSLSN SESSNSIADA ISSKSASVGP PTTAASSASS SASDSNSLAS NAPSPASPED1080
1081 CSAAPSPACS ASTTGSIPPS TVVDIAMVEA TSKSLPKSAI SPILSQPKTI RFPAGAGASG1140
1141 KGGKRHDGVC YWDKCNKKHE SNSKLLDHMQ THHVNTQTGP FACLWVGCKV YNKESCSRRW1200
1201 LERHVLSHGG SKQFKCIVEG CGLRFGSQLA LQKHVNNHFN ATDNARESTS KRTSDPPVPK1260
1261 QLRKNGKKLR YRRQPFSARM FDFFDTGIME GLQHRLRQIS TLTNGAQAIE FQGQCMMRRR1320
1321 NSQGGYECFV RWSPREIISD EWLPECLNRT RQHTKVLHIK QMRPAEKTRV DSLLSTAFRL1380
1381 RYDSHLFADD YNVNEQQVGE ASGSDCDEDG DGDQEEEDLE DQDEDEEEDG SSSSRSASCE1440
1441 TVSSYQQVLS IAKLQMQQRR KHPRKPPKVT PPAREKLVPT DALVPI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
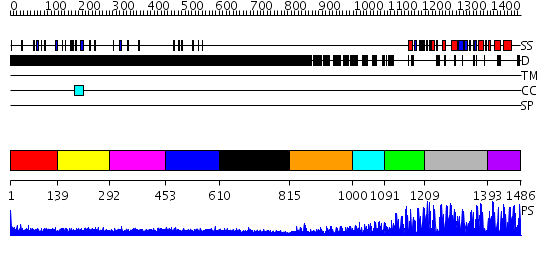
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] | 1.041981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [139..291] | 1.055982 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [292..452] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [453..609] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [610..814] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [815..999] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1000..1090] | 1.469995 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1091..1208] | 28.522879 | Transcription factor IIIA, TFIIIA | |
| 9 | View Details | [1209..1392] | 45.09691 | No description for 2i13A was found. | |
| 10 | View Details | [1393..1486] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 | No functions predicted. | ||||||
| 7 | No functions predicted. | ||||||
| 8 |
|
||||||
| 9 | No functions predicted. | ||||||
| 10 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|