













| General Information: |
|
| Name(s) found: |
CG14470-PA /
FBpp0085385
[FlyBase]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1976 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: |
serine-type endopeptidase inhibitor activity
[IEA]
endopeptidase inhibitor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMQEIQSIV ANGDGEGPSI IHPAPLAESH PDKNVQDIVN SVQNVLNKEM ASSSAKNSSG 60
61 ELLLPLLDTP DVSSSWSLIL PAIFGNTGGD IFPQQRPQQI VMTAESELLE TPNKTPTGTT 120
121 MATKKPKPAK RKPGTKRKTT TTPSTTAVTP QVVQEELDPM ESVYTGMQQY QQVAANMPHF 180
181 ELVHMQPIYQ KQPSSTSERP ETTTRRVFYN NQEIIHTPTS SSSAPSTTEV VLTHQLAKKP 240
241 ASVQRKPNQP HRVKKPNGNG TTQTTSQAGK QKIKRKTTTT STTTTTTTTP APEPETQPPT 300
301 QEEPVPATTE STTTTTQAPA STTPKKKKRK PTRTPGTGSS GGSSKPSKPK RKPTSKPRPQ 360
361 IQEVSGSQIV VPQKPSRPQR PTSNPSITST MPAQEVTTTT TTFKPVITST EPQTTEPPAE 420
421 STVTTTSEPE PTTTSSTQAS TTTSTETYTT TSTEAPTTTV TAPTTTSIAP AKTTAIPPAL 480
481 HHQKLHNKPA NRPLSQQHNH GLGRPVHYTT TESSKIVGEP QTETYGIVYV IHSSTTTHKP 540
541 FYPLPSYNDD AVQNVSTLLT PPPVQHKPLK KTPLPSLSQL QQQLHLGSPS PAPQLSPDQI 600
601 LSMDQIVQSL TQDLSASDKG EEASGASPVK YDSAESKLEI EALANKKKVA VVSSTTPAST 660
661 TTPSSSSITT TPSKPTKLST SHKPLTSHYG GSTLATMLSE EDEDITEFTE SSVVQQQPAD 720
721 TTISSLENEN EDQTPVVVIT ARPQYFTVTR VPEPLQHIKT DHRQEETETT ELPGLTQFLV 780
781 TTQMSQVDDD VDESTDKSVV ESVHENVSEA SQSYPTDLEV KEEVPAAVVT TEQTTKLPVS 840
841 ETSTVAEMDS TEVTPLIADL VQQLNLEMLK PTDQISMANF QTQAATVAST LVDGSNTLVS 900
901 DVPLNSNETK DELEFLISDV IQQITQGDQN KLPPNAQPAD DSNRLTSFAQ LNTSFLLQPN 960
961 IEKTEVNQIQ QTHGTTTAGP FKKESTYPEA LPESPTHLET STHRIPILSL SQIYAQQQQD1020
1021 EDEEQQVFAN ERIEVQSTME TIGQETVATT EEIYTETPSV ETTDADSVES TTPIEPQPQD1080
1081 PTQQTTSSEE GSGSEEPTTT YSNIPESNTN RENEEEILQL LKNDTKEQKQ TPNIDIQEKE1140
1141 TEPEPSTSEE PGSTTQLSVV EEITLPTNIY QDAQEEEQSS TTEKHVYLEP QPQEAQIVVE1200
1201 VTDTNAAQLQ PEYQYPPSGE REEDKSPVTQ IPGDMSLASS DSDMSLSSEQ VTEKIELSTV1260
1261 KSQQLNDESM EEISDELTAT ISEEQDAESA EPVTSTQEIQ LDNEDPYVKL GETTATVVRP1320
1321 LNPNSEGVGE NKSATESDED NYNYKVKLPL SNYGNPPVDQ ADEEDEDDQA AYHIPPMLAS1380
1381 STTVTTSSTK ITITEAPARS TSTTSVPPPT TKRLSTLKPV SYYVQPSLMG GYNQLDMQPP1440
1441 KVLPYNANKA DPVQLQVPIH SQSMLNQQRP SQRPPLLINL MASSTQSTRP PVKLDPSPES1500
1501 SQGLEASTAM MDEDLQSFAR LCNELAFSYW RAITSEKISS ARSLVISPFA LTSMLSMVFL1560
1561 GARGSTSGEM NEILKLDDMV TFNPHLIFKN ITNSVEQASD SDIATAAFVR EIFSDRANGK1620
1621 ILPFFKEKTQ QLYAGHVEEV NFHVVNDIVR RRTNLLVKRH TMGKVLEYLR TNSVWVNGPL1680
1681 ATISANLFQT DCSHGSTTDR DGEMFFQVHP TVRQRRLVPI PAVLYRSGFL AGYEPSLDAT1740
1741 VVSFGRVQNT VSTVYVMPGH QSSISPMDNL DRLERSLVET AFSDKQAWRR LLTSLMDRPG1800
1801 MEVQLPRFSH RSFVNASLGL QKMGLRGLFK SDFADLRGLT GAGNRDIFLS DMIQINTFST1860
1861 CGEEKISEHH HVEMYPAPPL RKRNKDVDAT DDDAFDSSER VVDFGSLVQE SALGRGFYDD1920
1921 LLDPKYLELP LPLRPRQARV PDAPRLRFDK PFLYFVRHNP TGMILFMGRF NPRLLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
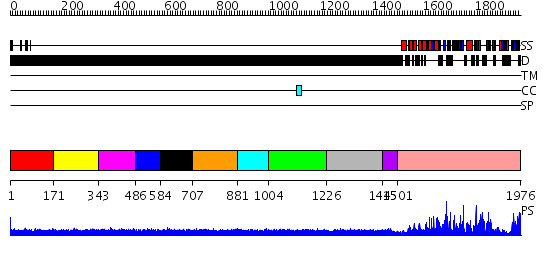
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [171..342] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [343..485] | 1.131986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [486..583] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [584..706] | 1.136984 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [707..880] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [881..1003] | 1.134985 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1004..1225] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1226..1444] | 1.112982 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1445..1500] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1501..1976] | 122.0 | No description for 2dutA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)