













| General Information: |
|
| Name(s) found: |
KCC2G_RAT
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Rattus norvegicus |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: |
calcium- and calmodulin-dependent protein kinase complex
[TAS]
cytosol [UNKNOWN] plasma membrane [UNKNOWN] endocytic vesicle membrane [UNKNOWN] nucleoplasm [UNKNOWN] |
| Biological Process: |
response to oxidative stress
[IDA]
response to hypoxia [IDA] regulation of long-term neuronal synaptic plasticity [TAS] G1/S transition of mitotic cell cycle [UNKNOWN][IEA] calcium ion transport [UNKNOWN][IEA] protein amino acid autophosphorylation [UNKNOWN][IDA] protein amino acid phosphorylation [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IEA] calmodulin binding [IEA] calmodulin-dependent protein kinase activity [IDA][IEA] protein serine/threonine kinase activity [UNKNOWN] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATTATCTRF TDDYQLFEEL GKGAFSVVRR CVKKTSTQEY AAKIINTKKL SARDHQKLER 60
61 EARICRLLKH PNIVRLHDSI SEEGFHYLVF DLVTGGELFE DIVAREYYSE ADASHCIHQI 120
121 LESVNHIHQH DIVHRDLKPE NLLLASKCKG AAVKLADFGL AIEVQGEQQA WFGFAGTPGY 180
181 LSPEVLRKDP YGKPVDIWAC GVILYILLVG YPPFWDEDQH KLYQQIKAGA YDFPSPEWDT 240
241 VTPEAKNLIN QMLTINPAKR ITADQALKHP WVCQRSTVAS MMHRQETVEC LRKFNARRKL 300
301 KGAILTTMLV SRNFSVGRQS SAPASPAASA AGLAGQAAKS LLNKKSDGGV KKRKSSSSVH 360
361 LMEPQTTVVH NATDGIKGST ESCNTTTEDE DLKVRKQEII KITEQLIEAI NNGDFEAYTK 420
421 ICDPGLTSFE PEALGNLVEG MDFHKFYFEN LLSKNSKPIH TTILNPHVHV IGEDAACIAY 480
481 IRLTQYIDGQ GRPRTSQSEE TRVWHRRDGK WLNVHYHCSG APAAPLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
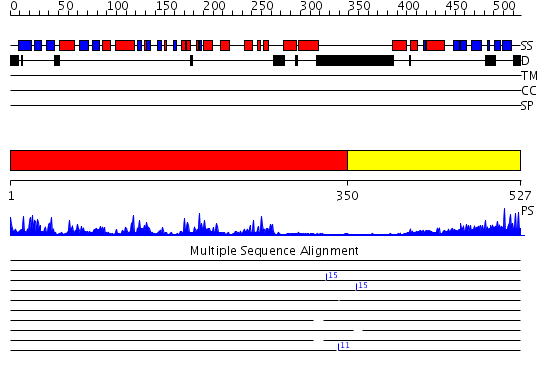
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..349] | 79.0 | No description for 2v7oA was found. | |
| 2 | View Details | [350..527] | 47.30103 | No description for 2ux0A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)