













| General Information: |
|
| Name(s) found: |
YC79_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 767 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHARMQPHED RSRSGMSRSE AMELEDQLEE EERGGNVVVG SSSANANAST AETGKKSAGG 60
61 SKPKRRRGER IPPSKRIRKA IAYVNKYASG CFGRVERRLV CTRSISRHSN QKLLDFCVEC 120
121 KKHKIKCVGN FPCGRCLKKG LECVIETPPR LLMNKDEISL NLQRLSHMET ILSKLMPSMS 180
181 LDLPSLEAKI AELSESLQTY PDKVLTDIEL GSYQQVTLNE TQTYYEDAGS NESFVARVHE 240
241 IICQGREFQV QHKLTNKGNK TFEDILDPAD NTVASLIHAL PPKDITFYLL MTFWQFSSDN 300
301 NHFYYNTKLF AAKVHHLFDD PTSFQSKDGG FVCMLLLSMA MGSLFSYIRH PEFLSDENHD 360
361 RWTYPGSQFY QNAKLLFPKV ISESSLETVQ SFFLAGMYLS PTLAHEVVYM YFGIAMRAAV 420
421 ANGMHKKSAN AQFSGDVAEL RKRLFWSVYC MERKIGISLG RPESLVRSEI DIHFPEYRES 480
481 LDSQNFIASF RTFTLAIKIS LLTNKVYDMW YSSLHGKANL KAATIKEIVN EIEAWRQQLS 540
541 PDLEIQNIGP DSRSYRGIVH LHLAYHIVRI AMGRPFLLHR LRERTMNSKT DEGARLLTDK 600
601 LISYCYSSAL HIVDLLVLLR MHKFLSVYSF MDYHSCHAAS FIILVHLLIN PSEQTIEQLN 660
661 TAIDILNFVT DRFPLLKGST DVITNLRAFA EQSEVYQSRL NATEPAYSTQ VPFFPRDADY 720
721 HNQINLQNLW NDENINASLE ALFNDAKGFG FLLPADNFIF PSDDGDM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
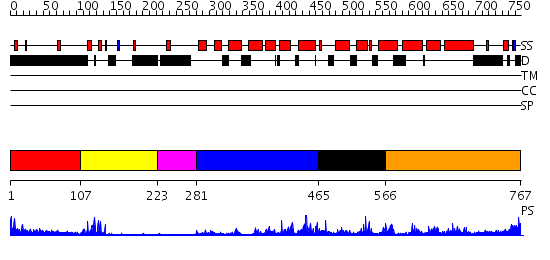
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..106] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [107..222] | 2.21 | PPR1 | |
| 3 | View Details | [223..280] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [281..464] | 3.362995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [465..565] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [566..767] | 2.261995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.36 |
Source: Reynolds et al. (2008)