













| General Information: |
|
| Name(s) found: |
YCZ8_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 967 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQNAPKCYLP NSRKGRDVNF HDRGPETLKC LYDESEDNNN FTMHSESIGP KSLDSLSPRR 60
61 LSNYSSAVDP RTTSLEDVPR LILTRSNSQE QTPLRTHTPV EYMSDSIHKS LADLQLGGSS 120
121 YSVAESPVPV LQSSAVSEAD DMASAHSAHP SRKASRSLRL FESSTSNNLE TGNSTNTALH 180
181 NVSSPLESVS ERLSKSGSPS VGAQHDLDEV ISEKDTSLSR RSSRGRSSAP KRRKDSGSSK 240
241 TTATYIPHNP SKKSSQHLPL LPDASFELER SVSRSYQSPA STPRSSVSSV SSSPPEDHPF 300
301 FHWNVDYPVT VRLEPFKHQV GGHTAFFRFS KRAVCKPLTR NENTFYETIE ACHPELLPFI 360
361 PKYIGVLNVT HTITKTEEND STTEYVNTES SSKTPAPHKH TFNSCYQKKD YGYIPEVSVE 420
421 QNRHIFPEWM LPDKRSHSYG SPKSLHHKSS SAGERPVSPT FVADIPPKTP WGTTLINRKL 480
481 REEVLREVFA PKHARRRLGT RFHSRSSHRP SVFRDNSVAF GQLDNGNTSS RARDKDADPN 540
541 KSLSCSVEDK HYDLHSAVAE ENEEVDEELL NVPSNNQGKS YRRFSSDAVW EEPESNEFPR 600
601 VSGTMEDYDS RESTGHTIKE LRSTPNSHGT VPDDSIFAMD NEENSELPPP LEPAEIGDPF 660
661 RSVNDPRRVL SLPHMASADE DHRIPASDNQ NNNNNDANAL AENSESQHST QIERYIVIED 720
721 LTSGMKRPCV LDVKMGTRQY GIMATEKKKA SQTKKCAMTT SRVLGVRICG MQVWHPWLQS 780
781 YTFEDKYVGR DIKAGEEFQH ALMRYLGKTD DDEDNSHLLV HHIPTIIRKL EQLEQIVRFL 840
841 KGSRLYASSL LFLYDGEPPP SDKSSKEKVK PREIDIRIVD FANCVFAEDK ELLAKATCPP 900
901 QHKDTYDRGY VRGLRTLRLY FLKIWKEAKG MQIAERGYED SLSNAYDELG GLMSYSNDDD 960
961 SCGETST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
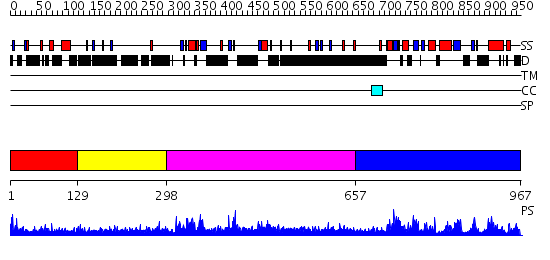
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 2.248996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [129..297] | 5.257994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [298..656] | 2.34 | No description for 2iewA was found. | |
| 4 | View Details | [657..967] | 59.522879 | CRYSTAL STRUCTURE OF THE CATALYTIC CORE OF INOSITOL 1,4,5-TRISPHOSPHATE 3-KINASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)