













| General Information: |
|
| Name(s) found: |
NOP2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 608 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRKQKSKQG IPPTLEENHN SSHKVTENAK KRKHSKEKPQ NSRKRQLAEE KKSLFENSDS 60
61 ENEKDLIDAD EFEEAETLSD LEHDEEPQTF ADEFIDDEAK ECEGEEEDSV FDSDEEHEVK 120
121 PMFSDDSGDE EDLELANMEA MSRKLDEEAE LEEKEAEEEL HTNIHPEAPT VLPPIDGFTD 180
181 SQPISTLPQD LSQIQLRIQE IVRVLNDFKN LCEPGRNRSE YVDQLLNDIC AYYGYSRFLA 240
241 EKLFELFSVS EAVEFFEANE MPRPVTIRTN TLKTQRRELA QALINRGVNL EPIGKWSKVG 300
301 LQVFESQVPI GATPEYLAGH YILQAASSFL PVMALAPQPN ERILDMSSAP GGKVTYVAAL 360
361 QKNTGIIFAN DSNKARTKAL SANIHRLGVR NAIVCNYDGR KFPNEVIGGF DRVLLDAPCS 420
421 GTGVIYKDQS VKTNKSERDF DTLSHLQRQL LLSAIDSVNA DSKTGGFIVY STCSITVDED 480
481 EAVIQYALKK RPNVKLVSTG LEFGREGFTR FREKRFHPSL KLTRRYYPHV HNIDGFFVAK 540
541 LKKISDKIPT VNVADDMKDG TNNDVEIEKN STEIDNITFN DEADKEIIEQ NRRKWLKSKG 600
601 YKVAKKKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
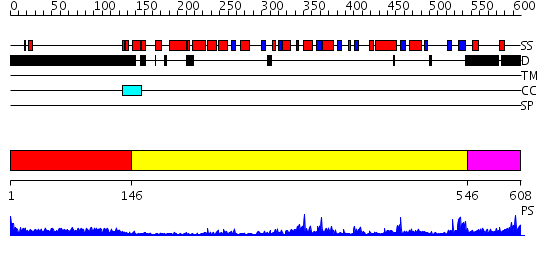
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..145] | 17.231997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [146..545] | 55.0 | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | |
| 3 | View Details | [546..608] | 62.045757 | No description for 2frxA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)