













| General Information: |
|
| Name(s) found: |
RSC9_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 780 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQVIQGAVE EPVYSNKSLT NENDSFLSLI ESFSQERGVP IDINPKIGRK PILLYELYKK 60
61 VIKRGGYDAV SATEDGWTNI AEEFNQSDPA RSAGILQNVY FKYLISWEIH DHWHLLLPPP 120
121 STLELSENRQ NVLERVKVFN SCSPPKPTNP ITIIDNDKTH SFKKFYKSLV KEDEENGIEK 180
181 KINEAMNSSA SQPLPNSTNF ASTTFPSVPF HPLPVDSGLQ KYIDRNSNLT PPALGAGVPG 240
241 PPLLVRVALA LKSQQDKEVD WAIGHLIKIS FERHQEFKLE RLPSLAECLV YSLGFQVTKA 300
301 KQVSDLTDIS LCMDRAIGIA LVLRNSVLSV ENAKHVAQTK LVISVLEASI RCAKTFNNLE 360
361 FLHYCLDISE MISSYLHVED EKNVFYLALC EFLNSSDYSI LIATLRTLAR LALNDRNNRL 420
421 LQDLKSNVIA NIIALVETDN EEIVAAALDF LYQYTTYRTN TSNLLASQEA WMLVDQLIRL 480
481 MMYQAQERFM TVTIENSESN PAVKHEATSS ISLPMEELQQ LVAMREPERS VKWMRCCFEP 540
541 SSDDYVLQTD IWQLYCSDME RAQGPNVMGI SPADFIKVSS HAFYNARAMT VSTPQLPVEY 600
601 VINGIKRRKF PTSVNGMRYQ PCRWCLDSGK ECGELLLGTP LLHSHLQEMH IFPQILETGK 660
661 CRWSDCKYEI QRLTPASELS HYQLLSHIVT HLHDDSLETL VEGRKLSPSR EFRIPLLLTA 720
721 VDDQGDATGI ALTATLVLRN LVRSKQGKML FSAIESRVLE VSSLNPAVAP YVSEMLLGQI 780
781 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
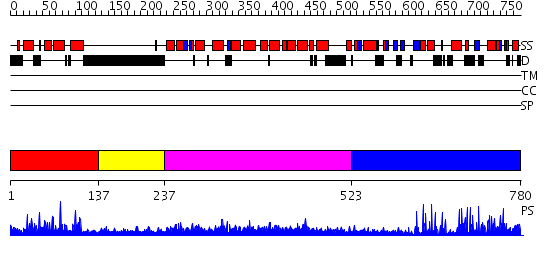
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..136] | 32.30103 | DNA-binding domain from the dead ringer protein | |
| 2 | View Details | [137..236] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [237..522] | 1.28 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP | |
| 4 | View Details | [523..780] | 39.69897 | No description for 2i13A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)