













| General Information: |
|
| Name(s) found: |
YME2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 773 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNLFLSRFTG LRNTSFLGHA RPFMFPRRYA HSLVAHNIGH IKLDPNEGLF FVDNLVNTPT 60
61 YFYIQRYTAL LFQNNLKKQL SAAFPSSDTM QLEDIIFRWQ DGGAFLKVRY KEFPQDTEAV 120
121 SEHVRESFRK RPVRTILHPF STPIPHMVHG IPWLQDLYIF PSRTVDVNFE GPPLSQERLY 180
181 SIFRTYGKLR SVTINSPTSA TLSFSSLRSA TSALNCMHGF VYGKTEFHMR YRHMNRFVAF 240
241 KDWLFSHPRF TIPLVAAAIT VLTASLFDPI RKFFVETNIV HGQKLRNINV VGWVKKKTRD 300
301 IVLSPFHENG TNIKAPIWTT REKDCEQLKE WLDEALHSFI VVQGPRGSGK RDLVDRVVKE 360
361 RKNVLFFDCD RLFSTTNTEM FVSTLASQTG YFPLFSFLNN ISSLIDMAAQ GLIGQKTGIV 420
421 SSSEGQVRQI LNTTQTVLRS LALREHKEAD TSVLDESEFL EVHADRLPVV ILDNFQLRKL 480
481 SNPMQRVVAE WAGNLVKEGI AHVLMLTPDV GGTKSLEQYV NGWENRTLLL GDADPVLAQR 540
541 YVIESLPEEM QTEELRKELR VQLPKIGGRL RDLDYVARRL RVNSSSVSEA IGGIVSQNAS 600
601 DILQTFLRPA SLTSEEKPTF TPEESWTLIT YLSQHEYIPY HMLMLDPLFK GHDDAVRALE 660
661 ESELITITTV NARPDKVYAG KPVYVTAFRQ LVNDPVLSAN MQLVRCNALI GMANNAIKND 720
721 EQELQMLKDL GNLESGVKDR AHYLTTRIQK NQGVITDNEK SIERLTEALK KID |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
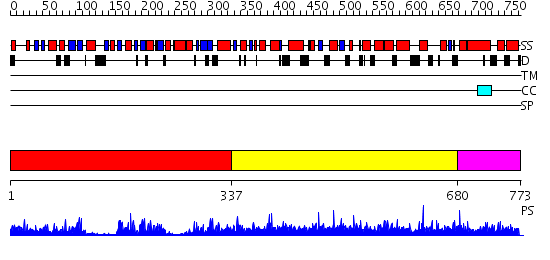
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..336] | 19.39794 | No description for 2ghpA was found. | |
| 2 | View Details | [337..679] | 1.01 | Crystal structure of (13814777) from SULFOLOBUS SOLFATARICUS at 2.00 A resolution | |
| 3 | View Details | [680..773] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.46 |
Source: Reynolds et al. (2008)