













| General Information: |
|
| Name(s) found: |
DCP2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 741 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFTNATFSQ VLDDLSARFI LNLPAEEQSS VERLCFQIEQ AHWFYEDFIR AQNDQLPSLG 60
61 LRVFSAKLFA HCPLLWKWSK VHEEAFDDFL RYKTRIPVRG AIMLDMSMQQ CVLVKGWKAS 120
121 SGWGFPKGKI DKDESDVDCA IREVYEETGF DCSSRINPNE FIDMTIRGQN VRLYIIPGIS 180
181 LDTRFESRTR KEISKIEWHN LMDLPTFKKN KPQTMKNKFY MVIPFLAPLK KWIKKRNIAN 240
241 NTTKEKNISV DVDADASSQL LSLLKSSTAP SDLATPQPST FPQPPVESHS SFDIKQKILH 300
301 LLNEGNEPKS PIQLPPVSNL PLNPPIQSSN SRLSHDNNSF DPFAYLGLDP KNPSASFPRV 360
361 VSQNNMLTNK PVLNNHFQQS MYSNLLKDQN SVQHLFAASD MPSPMELPSP STVYHQVFYP 420
421 PTSTSVSSYG LGKTPQPAYG SSSPYVNGHQ TQQISSLPPF QSQTQFLARN SDNSGQSYNS 480
481 EGDSNSKRLL SMLSQQDTTP SSSTLSKEAN VQLANLFLTP NSLETKKFSD NSQGEEISDN 540
541 LHGESCNNPN ANSVHSAQLL QALLHPSATE TKEETPKKTS DSLSLLTLLK SGLPTPANDL 600
601 QNKSQNNERK ASSQVKELEV KNYSKSTDLL KKTLRIPRND EPLEAANQFD LLKVSPQQKS 660
661 EVPPKRNELS QSKLKNRKKK ENSETNKNHV DMSPGFVKIL KRSPLADQKK EDTQESDFKG 720
721 SDDHFLSYLQ SVVSSNSNGL H |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
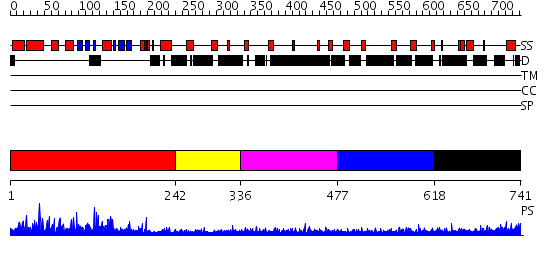
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..241] | 104.0 | Crystal structure of S.pombe mRNA decapping enzyme Dcp2p | |
| 2 | View Details | [242..335] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [336..476] | 2.270994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [477..617] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [618..741] | 2.248993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)