













| General Information: |
|
| Name(s) found: |
YK42_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNISSQNVLL PSPIPSSSPM ASHKKSWLSK HPNQSMTFEK PQLGQFVLTP EPSSNTFYAP 60
61 SSPASAVRRE PLSPMSFVRM RSHRVNKIGR SSQQCDHVLS TVDKAISRVH AIVTCTQDRM 120
121 IIECVGWNGM IVSDKMRKSV FHMKKNDRIV LVRPNSDACP VLDVFGYRVL LGWPSDSEDE 180
181 WEGNLNAKNY EENREPMSPS PQEALPLMPS SPPSQDYQND QNHLILYTNS ESIPKLNLRS 240
241 NELVYPPPSK DLLQKLLALE KDGQVEKSDC SKNTQLKPSF LPKNTDDLLN GTDDNNIVLR 300
301 EVKVSFENEK IESDDLDKNE EISEGEEYTP IEESKEPITV RRDSVIQIDE SSAGLTDVIS 360
361 ELNFTNHNDD SKNSNITTSN DSPVNEVEPM APELSSAVVE KKEPEDYESI SAVDENTNDS 420
421 NESLPSSHDY SESTKENSAP DSLLLGLVLD ELVFSTTSTT PLPALSHLFP SNMPLQLIQD 480
481 KLRDLAAKHP YFEEVKRYGT DANGDPLWSE WFYNPDVDDD LERRMRYAPL MRPVRSSRRV 540
541 HKQYYWKKPR ARPRSSGHSS RRRRLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
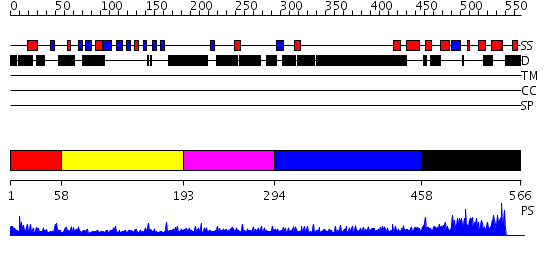
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..57] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [58..192] | 1.34 | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | |
| 3 | View Details | [193..293] | 4.199997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [294..457] | 1.191994 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [458..566] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)