













| General Information: |
|
| Name(s) found: |
VAM6_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 905 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHRAFSLYRV LELSKARVEC VFELGGLVYV SNSNGDLDSY KIYNNEEEEA ADFVMEHVDV 60
61 YPNFTKKPIT KVVSCATQDI FYALSDSQVY VYQISTFKKL FSFGAHCQNM CLYGDELIVL 120
121 SSKKNLEIYE IQKNSKPNLT KTISLNDRPR SLAWVSPTMI LVSLSNDFCA VNTETSRISS 180
181 LNLAWQQSSS LGLGISYIGM SIKSNKLHIT RISDDEVLLS KDSQGLLVNL KSLQVSRNPL 240
241 RWPTVPQAVI YNSPYIITLH NQYIYIWNKE TYAMIQQIGI SNIYSTFSCH KNTFFTSNSY 300
301 VWILTPEDFS NQIEALLNTE NLNEAISVLS QITVSQFPKR DYYLRITKRE KALRSFSSGD 360
361 YDLAMRLFSE ISESPSTVLG LFPGLLDNNY SDAISILSMA PSQNESIESN VLFPGNHSNS 420
421 QTDLRNGDAV STVANNKRLR SLSTYLTDSR RKANRFLSYD EEHYFLQKKN LFLNADGTLV 480
481 AKEKLEKIAV QIDTTLFLIY MISSPALVGS LLRLPNRCET SVVETNLLSA KMYRELVEYY 540
541 YGKSLHEAAL DLLTKLCDEP TDTLSLKGKS NTTSKYEPIL SYLEKLSPEL DHLIFKYSRV 600
601 PLSEDPQNSI VIFIDENSEA STISKGVVLK YLETISYKVS IIYLEKLLLD NKFNDTVFPT 660
661 RLALLYLKRI LELEETTDFK NQEVFKQTIE KLEDYLTNSK QYDANVVLQE INSQDEFLST 720
721 VSIILYRRLS RHQDALDVYL KILNDWEGAL SYCNSVYSID GETEPYYMLL AEISKNYKSG 780
781 SLNILDFITK YSSRLDLNRV FPLLPKNISM KSYHSLFSSQ FRQLFEELSN KETQSKLYQK 840
841 RLEDLNEELT KVRSEKVVIT REKTCLFCHK RLGKSVISIF PDGSVVHYGC AKKYVSSNHL 900
901 PYEAY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
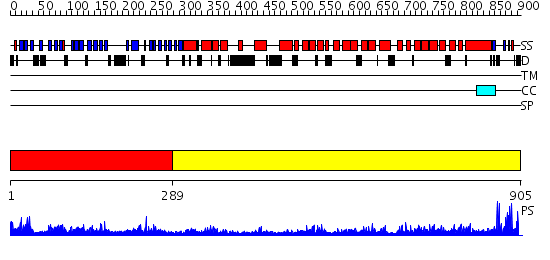
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..288] | 1.24 | No description for 2i3sA was found. | |
| 2 | View Details | [289..905] | 77.0 | Clathrin D6 Coat |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)