













| General Information: |
|
| Name(s) found: |
SCD1_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 872 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAYFQDRKTS SRSLPSYINH STQNLVGPRK DETNLSEYMK LRLLQSPSQV IYNLENTVSL 60
61 YRRCLNLRKR LMDISELAAF FDSIHREALN SSFKILEFKD IEFDDPVTEI WLFCRLGYPL 120
121 CALFNCLPVK QKLEVNSSVS LENTNVCKAS LYRFMLMCKN ELGLTDAALF SISEIYKPST 180
181 APLVRALQTI ELLLKKYEVS NTTKSSSTPS PSTDDNVPTG TLNSLIASGR RVTAELYETE 240
241 LKYIQDLEYL SNYMVILQQK QILSQDTILS IFTNLNEILD FQRRFLVGLE MNLSLPVEEQ 300
301 RLGALFIALE EGFSVYQVFC TNFPNAQQLI IDNQNQLLKV ANLLEPSYEL PALLIKPIQR 360
361 ICKYPLLLNQ LLKGTPSGYQ YEEELKQGMA CVVRVANQVN ETRRIHENRN AIIELEQRVI 420
421 DWKGYSLQYF GQLLVWDVVN VCKADIEREY HVYLFEKILL CCKEMSTLKR QARSISMNKK 480
481 TKRLDSLQLK GRILTSNITT VVPNHHMGSY AIQIFWRGDP QHESFILKLR NEESHKLWMS 540
541 VLNRLLWKNE HGSPKDIRSA ASTPANPVYN RSSSQTSKGY NSSDYDLLRT HSLDENVNSP 600
601 TSISSPSSKS SPFTKTTSKD TKSATTTDER PSDFIRLNSE ESVGTSSLRT SQTTSTIVSN 660
661 DSSSTASIPS QISRISQVNS LLNDYNYNRQ SHITRVYSGT DDGSSVSIFE DTSSSTKQKI 720
721 FDQPTTNDCD VMRPRQYSYS AGMKSDGSLL PSTKHTSLSS SSTSTSLSVR NTTNVKIRLR 780
781 LHEVSLVLVV AHDITFDELL AKVEHKIKLC GILKQAVPFR VRLKYVDEDG DFITITSDED 840
841 VLMAFETCTF ELMDPVHNKG MDTVSLHVVV YF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
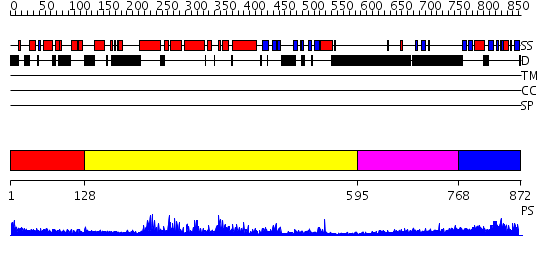
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 13.30103 | No description for 2d86A was found. | |
| 2 | View Details | [128..594] | 85.39794 | No description for 2pz1A was found. | |
| 3 | View Details | [595..767] | 1.533996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [768..872] | 2.58 | Solution Structure of the PB1 Domain of Cdc24p (Long Form) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)