













| General Information: |
|
| Name(s) found: |
DNAA_YERPN
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Yersinia pestis Nepal516 |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSLWQQCL ARLQDELPAT EFSMWIRPLQ AELSDNTLAL YAPNRFVLDW VRDKYLNNIN 60
61 GLLNDFCGTE VPLLRFEVGS KPAARAHNNP VTASVSAPVA PVTRSAPMRP SWDNSPAQPE 120
121 LSYRSNVNPK HTFDNFVEGK SNQLARAAAR QVADNPGGAY NPLFLYGGTG LGKTHLLHAV 180
181 GNGIMARKAN AKVVYMHSER FVQDMVKALQ NNAIEEFKRY YRSVDALLID DIQFFANKER 240
241 SQEEFFHTFN ALLEGNQQII LTSDRYPKEI NGVEDRLKSR FGWGLTVAIE PPELETRVAI 300
301 LMKKADENDI RLPGEVAFFI AKRLRSNVRE LEGALNRVIA NANFTGRAIT IDFVREALRD 360
361 LLALQEKLVT IDNIQKTVAE YYKIKVADLL SKRRSRSVAR PRQMAMALAK ELTNHSLPEI 420
421 GDAFGGRDHT TVLHACRKIE QLREESHDIK EDFSNLIRTL SS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
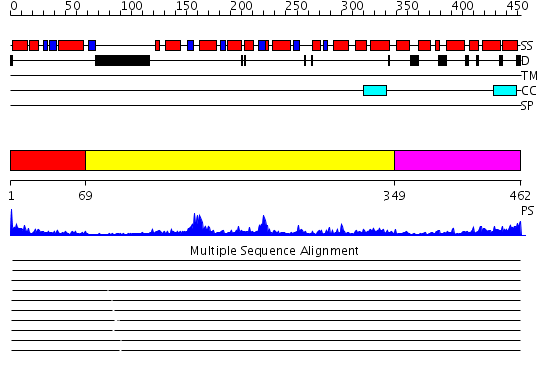
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 11.0 | No description for 2e0gA was found. | |
| 2 | View Details | [69..348] | 15.69897 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [349..462] | 32.0 | No description for 2hcbA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)