













| General Information: |
|
| Name(s) found: |
ASH2L
[HGNC (HUGO)]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAAGAGPGQ EAGAGPGPGA VANATGAEEG EMKPVAAGAA APPGEGISAA PTVEPSSGEA 60
61 EGGEANLVDV SGGLETESSN GKDTLEGAGD TSEVMDTQAG SVDEENGRQL GEVELQCGIC 120
121 TKWFTADTFG IDTSSCLPFM TNYSFHCNVC HHSGNTYFLR KQANLKEMCL SALANLTWQS 180
181 RTQDEHPKTM FSKDKDIIPF IDKYWECMTT RQRPGKMTWP NNIVKTMSKE RDVFLVKEHP 240
241 DPGSKDPEED YPKFGLLDQD LSNIGPAYDN QKQSSAVSTS GNLNGGIAAG SSGKGRGAKR 300
301 KQQDGGTTGT TKKARSDPLF SAQRLPPHGY PLEHPFNKDG YRYILAEPDP HAPDPEKLEL 360
361 DCWAGKPIPG DLYRACLYER VLLALHDRAP QLKISDDRLT VVGEKGYSMV RASHGVRKGA 420
421 WYFEITVDEM PPDTAARLGW SQPLGNLQAP LGYDKFSYSW RSKKGTKFHQ SIGKHYSSGY 480
481 GQGDVLGFYI NLPEDTETAK SLPDTYKDKA LIKFKSYLYF EEKDFVDKAE KSLKQTPHSE 540
541 IIFYKNGVNQ GVAYKDIFEG VYFPAISLYK SCTVSINFGP CFKYPPKDLT YRPMSDMGWG 600
601 AVVEHTLADV LYHVETEVDG RRSPPWEP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
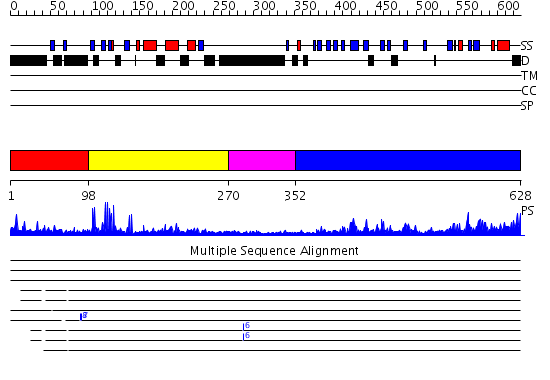
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [98..269] | 17.522879 | No description for 2f6jA was found. | |
| 3 | View Details | [270..351] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [352..628] | 32.0 | No description for 2fnjA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)