













| General Information: |
|
| Name(s) found: |
gi|215694337
[NCBI NR]
gi|115435848 [NCBI NR] gi|113532213 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 764 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFLQLADYHL RMDSRSPKIP RRSPDSKDKD SDRNKERDAK NDWDSSRAYG SETDCKEEMC 60
61 DSNKRKGLTM GEIVVDNSRS VDSCHETELH VLRDDRQDKS VEIKDILHDG VAKSDYAQRQ 120
121 IDLDSERRNG TGDNSRVDVL RDDKLDSGRD RNWSDRTREP EGSKDYVRNR QWQDSKEAND 180
181 SEWKNAHERL DGGSFHGRAG YRRDSRGRSE SIRGSSTYGG RYDSSDSIEI RPNNNLDFGR 240
241 EGSVSGRRYD VGAHRDATPG TNGDKSANPE ADQSGSTSTI SQFPQHGPKG DRSSRGRGRP 300
301 NSRDSQRVGG TLPIMPPPFG PLGLPPGPMQ HIGPNIPHSP GPLLPGVFVP PFPGPLVWPG 360
361 ARGVDVNMLS VPPNLPIPPV AAEHRFAPSM GAGPGHNIHL NQIGSGIGAP TNVSGLSFHQ 420
421 LGTQSREMAH DKPPAGGGWT PHRNSGPTRK APSRGEQNDY SQNFVDTGMR PQNFIRELDL 480
481 TSVAEDYPKL RELIQRKDEI VANSASPPMY YKCDLRQHVL SPEFFGTKFD VILVDPPWEE 540
541 YVHRAPGITD HIEYWNAEEI MNLKIEAIAD TPSFVFLWVG DGVGLEQGRQ CLKKWGFRRC 600
601 EDVCWVKTNK KNATPSLRHD SHTILQHSKE HCLMGIKGTV RRSTDGHVIH ANIDTDIIIA 660
661 EEPTDGSTKK PEDMYRIIEH FALGKRRLEL FGEDHNIRPG WLTLGKGLSY SNFNKEAYIK 720
721 NFADKDGKVW QGGGGRNPPP EAPHLVVTTP EIEGLRPKSP PQKN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
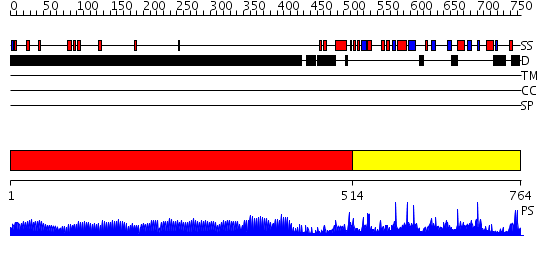
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..513] | 47.69897 | No description for 1y0fB was found. | |
| 2 | View Details | [514..764] | 2.32 | m.RsrI N6 adenosine-specific DNA methyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)