













| General Information: |
|
| Name(s) found: |
DNLI4_SCHPO
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 923 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDEAKETVFT KPQNHSSTLE FYDFVTTLLE PLSRIGKTRK SKTSNLDPYE LKRKILLDYF 60
61 NKWRQHVGPD LYPLLRLSNL IFLDRERGSY GFKEFGLGKL FIRAMHLSPT SEDAKSLKNW 120
121 RGSESKHTGD FSTMLQDILQ RRAYRTFPGA FTVGDVNALL DQLADASSEY VLNFLPYLTL 180
181 IRDTRVNILE QFYRSLSPLE LRWLIPILLK VRKYGTSEKF ILSVFHPDAA RLYRLCSSLK 240
241 RICWELYDPS RSLDETETDV EVFSCFQPQL ANFKKKDLHQ TLEAMGNKPF WIEEKLDGER 300
301 IQLHMSSGKF QFYSRNARSY TYAYGSSYFD EQSRLTQYII GAFDKRISQI ILDGEMVTWD 360
361 PVLETVIPYG SLRSIFEDSS SHSSYSPYYV VFDILYLNGK SLVKYSLESR RRILEKVIVR 420
421 ESHRMSILPY KVGSTIEDIE AELRNVIQEG SEGLVIKKPS GSYHLGERMD DWIKVKPYYL 480
481 QGFGEDLDCL ILGGYFGRGK QSGKINSFLC GLRMDYTPKD HSEKFQSFVR VGGGFTYFDR 540
541 DIIRKETEGK WLPWSSDALE YMELAGTKQD FEKPDMWIHP KDSLVLQIKA AEVVVSNRFK 600
601 TNYTLRFPRL EKVRLDRSWK DALTINEFFT LKNAVEKQDN VSFHVNKKRK VSQKREKQKK 660
661 FLYDEPTFKK EASPHSDVLK NLHFVVLPPT ELHETKAGLQ QIIIENGGLI HQGVGNFGKE 720
721 RLFLVADRVS TRVSIERSKN MCTIIRSQWV MDSVNNQRLM PQWSYLLFSK DEKYSWKTAL 780
781 ESLSAKSLSN LLVELKQLDL SKEYSKISDD TSILNLTISK EEASFVGAFP FLKFTVFLDL 840
841 KGIENSELYD VRMGQYRLTK CILLWNGATI EKDISSKKLT HVVMFVEDST RLEQLTKACE 900
901 LTQHRKFTQP SYFQRYGGSF CIP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
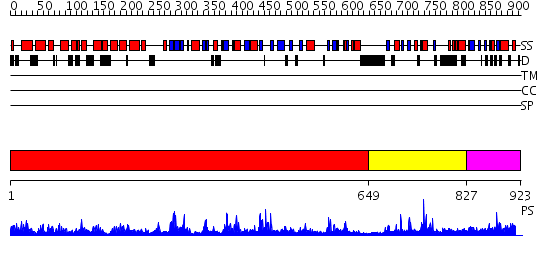
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..648] | 136.0 | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA | |
| 2 | View Details | [649..826] | 18.522879 | Co-Crystal Structure of Lif1p-Lig4p | |
| 3 | View Details | [827..923] | 17.154902 | The third BRCA1 C-terminus (BRCT) domain of Similar to S.pombe rad4+/cut5+ product |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)