













| General Information: |
|
| Name(s) found: |
CE40355
[WormBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 789 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP positive regulation of growth rate [IMP oligosaccharide metabolic process [IEA |
| Molecular Function: |
mannosyl-oligosaccharide glucosidase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHQPSRRRRP PREVERPSAT IRYEPVAEPE PWCSFCSWDL ILILLVMLGA GCFILLHLYL 60
61 YPNLEKVAPL PNIDPENAPY TWGTYRPHMY FGLRTRSPMS PLFGMMWYEQ PNTIQRPHIR 120
121 HWCNQDDRLP GYYWYEADGR HFGKQNISEA HKGVIQTDWI NDANGFAARV KLNMAPGRRY 180
181 NVILYLSAQE IGTRFRLGKH LSDVFHGYNE LLGKFTMSLR LKDNTKLQTS HSVMLTDEKI 240
241 PIDRYHDFVV DNTQAYNAPN QPLNYILNEK HNDEEGKFIA VQLNLGSQAE FDIILQTEKL 300
301 KGMKPEEFTN ILRIRSYNFN KKYENVFQLA GKNYTKTQLK MAKVSLSNML GSVGYWYGHN 360
361 RVLFNGIVQP YGPHVLFSAV PSRPFFPRGF LWDEGFHQML IRKMDSKMTL EAIASWMNAM 420
421 DTSGWIPREM IVGSEAEAKV PAEFIPQKND VANPPTLFYV MDKLVNDEKT VGRYAGILKL 480
481 LYPRLEKWFH WIRITQSGPT RTTYRWRGRN ETIKTELNPK TLSSGLDDFP RASHPSDLEY 540
541 HLDLRCWLAL ASRVLNRLAK SYGTDADYQR TAKAMEELNN FDSLVKDHWS EEAQGFFDYG 600
601 KHSFDVALSP VPTPGSPRQF EYQRVTSRAP SYTLVSDAFG YNNLFPMMLK LIPSKSPILK 660
661 SMLDKIRDPK ILWTNYGLRS ISRSSPYYMA RNTEHDPPYW RGYIWINVNY MVLSSLRHYA 720
721 DQPGPYRENA ENIFSELRAN LVKNLATQFQ KTGFLWENYD DRTGEGRGCH PFTGWSSLIL 780
781 LIMSDNLDT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
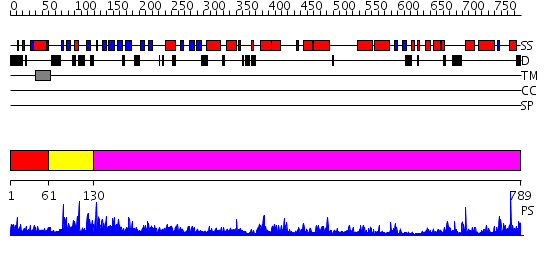
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 1.023997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [61..129] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [130..789] | 38.69897 | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|