













| General Information: |
|
| Name(s) found: |
CE40165
[WormBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 609 amino acids |
Gene Ontology: |
|
| Cellular Component: |
glycerol-3-phosphate dehydrogenase complex
[IEA |
| Biological Process: |
glycerol-3-phosphate metabolic process
[IEA electron transport [IEA |
| Molecular Function: |
oxidoreductase activity
[IEA glycerol-3-phosphate dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISRLLLQKS GILALGATAA STVAVWQTRT GFSKIREAPT GSTPGSLPTR ESIIESLKTE 60
61 KRFDVLVIGG GSAGAGVALD AQTRGLKTAM VEYGDYCSGT SSKSSKLLHG GVKYLETALK 120
121 EFDYEHYQIV QEGLNERLNI MKSAPFLSQT FPVLVPTYKW WQKIYYWGGV KVYDFLAGKN 180
181 VLKPSKFVSK EEAIEICPTI RQEGLKGAML YYDGQGNDAR LVLVVALTAI RNGAKCVNHT 240
241 ECIGLLKDSY GKVNGALVKD HISGETYEIH SKVVVNATGP FNDHIREMAD ETRNKIIVGS 300
301 SGIHLTVAKY FCPGNAGLIV PKSSDGRVIF AFPWENVTIV GTTDDPTEPS HSPTVTEKEI 360
361 QYILKEMNRA FEKEYEIKRE DVTSIWSGIR GLVQDPKRQK EHNSLARGHL VDVGPTGLIT 420
421 IAGGKLTTFR HMAEETVNKL LDINKILEAQ PCVTRGMMFE GAHNYTSMLY RTISQNYGIE 480
481 EQVATHLCQT YGDKVYEVLK LCKSTGQKFP VIGHRLHPDF PFLEAEVRYA VKEYARIPAD 540
541 ILARRTRLSL LDARAARQVL PRIVAIMAEE LKWSPSEQIF YFDEGMKFLK VENGSVQSWK 600
601 KEHSGVELP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
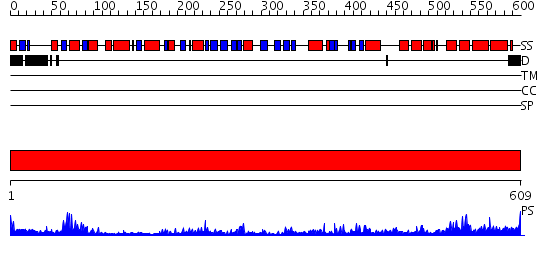
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..609] | 41.221849 | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|