













| General Information: |
|
| Name(s) found: |
SLC26A8
[HGNC (HUGO)]
|
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 970 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAQLERSAIS GFSSKSRRNS FAYDVKREVY NEETFQQEHK RKASSSGNMN INITTFRHHV 60
61 QCRCSWHRFL RCVLTIFPFL EWMCMYRLKD WLLGDLLAGI SVGLVQVPQG LTLSLLARQL 120
121 IPPLNIAYAA FCSSVIYVIF GSCHQMSIGS FFLVSALLIN VLKVSPFNNG QLVMGSFVKN 180
181 EFSAPSYLMG YNKSLSVVAT TTFLTGIIQL IMGVLGLGFI ATYLPESAMS AYLAAVALHI 240
241 MLSQLTFIFG IMISFHAGPI SFFYDIINYC VALPKANSTS ILVFLTVVVA LRINKCIRIS 300
301 FNQYPIEFPM ELFLIIGFTV IANKISMATE TSQTLIDMIP YSFLLPVTPD FSLLPKIILQ 360
361 AFSLSLVSSF LLIFLGKKIA SLHNYSVNSN QDLIAIGLCN VVSSFFRSCV FTGAIARTII 420
421 QDKSGGRQQF ASLVGAGVML LLMVKMGHFF YTLPNAVLAG IILSNVIPYL ETISNLPSLW 480
481 RQDQYDCALW MMTFSSSIFL GLDIGLIISV VSAFFITTVR SHRAKILLLG QIPNTNIYRS 540
541 INDYREIITI PGVKIFQCCS SITFVNVYYL KHKLLKEVDM VKVPLKEEEI FSLFNSSDTN 600
601 LQGGKICRCF CNCDDLEPLP RILYTERFEN KLDPEASSIN LIHCSHFESM NTSQTASEDQ 660
661 VPYTVSSVSQ KNQGQQYEEV EEVWLPNNSS RNSSPGLPDV AESQGRRSLI PYSDASLLPS 720
721 VHTIILDFSM VHYVDSRGLV VLRQICNAFQ NANILILIAG CHSSIVRAFE RNDFFDAGIT 780
781 KTQLFLSVHD AVLFALSRKV IGSSELSIDE SETVIRETYS ETDKNDNSRY KMSSSFLGSQ 840
841 KNVSPGFIKI QQPVEEESEL DLELESEQEA GLGLDLDLDR ELEPEMEPKA ETETKTQTEM 900
901 EPQPETEPEM EPNPKSRPRA HTFPQQRYWP MYHPSMASTQ SQTQTRTWSV ERRRHPMDSY 960
961 SPEGNSNEDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
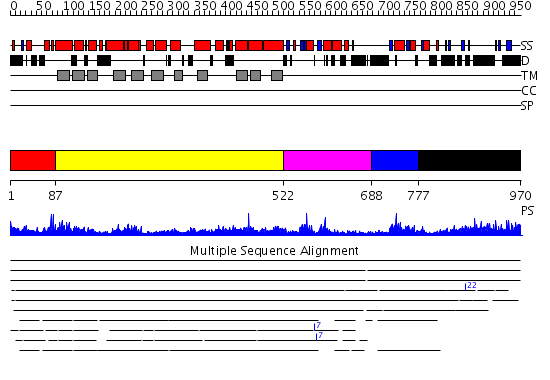
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [87..521] | 15.69897 | Cytochrome O ubiquinol oxidase, subunit I | |
| 3 | View Details | [522..687] | 2.42 | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES | |
| 4 | View Details | [688..776] | 4.045757 | Solution Structure of putative anti sigma factor antagonist from Thermotoga maritima (TM1442) | |
| 5 | View Details | [777..970] | 78.166997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|