













| General Information: |
|
| Name(s) found: |
ATG13 /
YPR185W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 738 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to membrane
[IDA |
| Biological Process: |
autophagy
[IMP protein targeting to vacuole [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAEEDIEKQ VLQLIDSFFL KTTLLICSTE SSRYQSSTEN IFLFDDTWFE DHSELVSELP 60
61 EIISKWSHYD GRKELPPLVV ETYLDLRQLN SSHLVRLKDH EGHLWNVCKG TKKQEIVMER 120
121 WLIELDNSSP TFKSYSEDET DVNELSKQLV LLFRYLLTLI QLLPTTELYQ LLIKSYNGPQ 180
181 NEGSSNPITS TGPLVSIRTC VLDGSKPILS KGRIGLSKPI INTYSNALNE SNLPAHLDQK 240
241 KITPVWTKFG LLRVSVSYRR DWKFEINNTN DELFSARHAS VSHNSQGPQN QPEQEGQSDQ 300
301 DIGKRQPQFQ QQQQPQQQQQ QQQQQQRQHQ VQTQQQRQIP DRRSLSLSPC TRANSFEPQS 360
361 WQKKVYPISR PVQPFKVGSI GSQSASRNPS NSSFFNQPPV HRPSMSSNYG PQMNIEGTSV 420
421 GSTSKYSSSF GNIRRHSSVK TTENAEKVSK AVKSPLQPQE SQEDLMDFVK LLEEKPDLTI 480
481 KKTSGNNPPN INISDSLIRY QNLKPSNDLL SEDLSVSLSM DPNHTYHRGR SDSHSPLPSI 540
541 SPSMHYGSLN SRMSQGANAS HLIARGGGNS STSALNSRRN SLDKSSNKQG MSGLPPIFGG 600
601 ESTSYHHDNK IQKYNQLGVE EDDDDENDRL LNQMGNSATK FKSSISPRSI DSISSSFIKS 660
661 RIPIRQPYHY SQPTTAPFQA QAKFHKPANK LIDNGNRSNS NNNNHNGNDA VGVMHNDEDD 720
721 QDDDLVFFMS DMNLSKEG |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
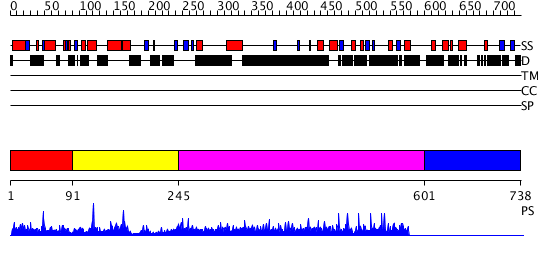
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [91..244] | 1.017991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [245..600] | 11.038996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [601..738] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)