













| General Information: |
|
| Name(s) found: |
STD1 /
YOR047C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 444 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA plasma membrane [IDA |
| Biological Process: |
response to salt stress
[IMP signal transduction [IMP glucose metabolic process [IMP regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
protein kinase activator activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFVSPPPATA RNQVLGKRKS KRHDENPKNV QPNADTEMTN SVPSIGFNSN LPHNNQEINT 60
61 PNHYNLSSNS GNVRSNNNFV TTPPEYADRA RIEIIKRLLP TAGTKPMEVN SNTAENANIQ 120
121 HINTPDSQSF VSDHSSSYES SIFSQPSTAL TDITTGSSLI DTKTPKFVTE VTLEDALPKT 180
181 FYDMYSPEVL MSDPANILYN GRPKFTKREL LDWDLNDIRS LLIVEQLRPE WGSQLPTVVT 240
241 SGINLPQFRL QLLPLSSSDE FIIATLVNSD LYIEANLDRN FKLTSAKYTV ASARKRHEEM 300
301 TGSKEPIMRL SKPEWRNIIE NYLLNVAVEA QCRYDFKQKR SEYKRWKLLN SNLKRPDMPP 360
361 PSLIPHGFKI HDCTNSGSLL KKALMKNLQL KNYKNDAKTL GAGTQKNVVN KVSLTSEERA 420
421 AIWFQCQTQV YQRLGLDWKP DGMS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
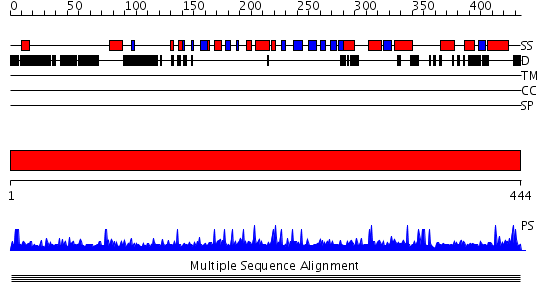
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..444] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)