













| General Information: |
|
| Name(s) found: |
KKQ8 /
YKL168C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 734 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
protein kinase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMDSKTLTAS MVMQEEKKRQ QPVTRRVRSF SESFKNLFRP PRSRDSSPIN VTRIPYRSSS 60
61 TSPKRSSEPP RRSTVSAQIL DPKNSPIRQR SYTLKCCTPG LSHPFRQTGS GASNSPTRHR 120
121 SISGEEQEIV NSLPEYKRSA SHTFHGIRRP RSRSSSVSSC DSSNGTTSSS DSQWAMDSLL 180
181 DDSDNDLTPY RGSNKDILKS KDRAPYNYID DYNKKALRRA TSYPNPLPSK QFYNERLYTR 240
241 RSHPDEESLE SLPRFAGADV QCIIEQNGFK VYEDGSHEHN IKLSGVIAKL EKGNSLPAHR 300
301 QGSLSRPRLG ITLSGLFKHH KNECDIENAL SLLPNVEKSQ TNHEKRTGQS PNDSNRSSPT 360
361 QGREDYLKIV NPDASLGSDE LKLINSLSSR IHKSLQNYLQ EKNLKPAECI GEQAPTFQDN 420
421 YGHPVGLVGA GAYGEVKLCA RLRNEKDSPP FETYHDSKYI YYAVKELKPK PDSDLEKFCT 480
481 KITSEFIIGH SLSHYHKNGK KPAPNILNVF DILEDSSSFI EVMEFCPAGD LYGMLVGKSK 540
541 LKGRLHPLEA DCFMKQLLHG VKFMHDHGIA HCDLKPENIL FYPHGLLKIC DFGTSSVFQT 600
601 AWERRVHAQK GIIGSEPYVA PEEFVDGEYY DPRLIDCWSC GVVYITMILG HYLWKVASRE 660
661 KDMSYDEFYK EMQRKNQFRV FEELKHVNSE LATNRKIALY RIFQWEPRKR ISVGKLLDMQ 720
721 WMKSTNCCLI YDST |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
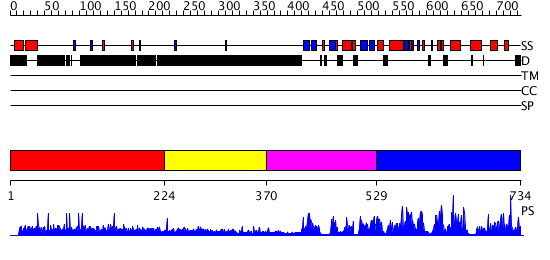
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 8.91 | No description for 1ldvA was found. | |
| 2 | View Details | [224..369] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [370..528] | 260.035458 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 4 | View Details | [529..734] | 260.035458 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)